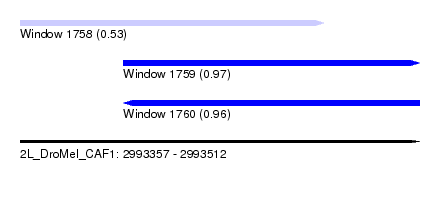
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,993,357 – 2,993,512 |
| Length | 155 |
| Max. P | 0.973052 |

| Location | 2,993,357 – 2,993,475 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.45 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -25.35 |
| Energy contribution | -27.22 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2993357 118 + 22407834 AUCAAAUCUAAAUUCAUCAGGUGAGCGAGCAGCAGCAGCAACAACAACCACAGUGAGUGCCCAAAUAGGUGGGCACACGUACACAGGUGUGUGUG--UGGCUGUGUGCGUAAGUGUGUCA .............((((...))))((..((....)).)).........((((.(..(((((((......)))))))((((((((((.(.......--.).)))))))))).).))))... ( -36.00) >DroSec_CAF1 5126 107 + 1 AUCAAUUCCAAAUUCAUCAGGUGAGCGAGCAGC---AGAAACAACAACCACAGUGAGUGCCCAAAUAGGUGGGCACACGUACACAGGUAUGUGU----------GUGCGUAAGUGUGUCA .((..........((((...))))((.....))---.)).........((((.(..(((((((......)))))))(((((((((......)))----------)))))).).))))... ( -31.20) >DroSim_CAF1 5100 115 + 1 AUCAAAUCCAAAUUCAUCAGGUGAGCGAGCAGC---AGCAACAACAACCACAGAGAGUGCCCAAAUAGGUGGGCACACGUACACAGGUGUGUGUG--UGGCUGUGUGCGUAAGUGUGUCA ....................((..((.....))---.)).........((((....(((((((......)))))))((((((((((.(.......--.).))))))))))...))))... ( -34.20) >DroYak_CAF1 5128 118 + 1 AUCAAAUCCAAAUUCAUCAGGUGAGCGAGCACCAGCAGCAACAACAACCACAC--AGUGCCCAAAUAGGUGGGCACACAUACACAGGUGUGUGUGAGCGGGUGUGUGCGUAAGUGUGUCA ...................((((......))))....(((.((...(((((((--((((((((......))))))).(((((((....)))))))......)))))).))...))))).. ( -38.40) >consensus AUCAAAUCCAAAUUCAUCAGGUGAGCGAGCAGC___AGCAACAACAACCACAGUGAGUGCCCAAAUAGGUGGGCACACGUACACAGGUGUGUGUG__UGGCUGUGUGCGUAAGUGUGUCA ............(((((...))))).....................(((((.....(((((((......)))))))((((((((((.((........)).))))))))))..))).)).. (-25.35 = -27.22 + 1.87)

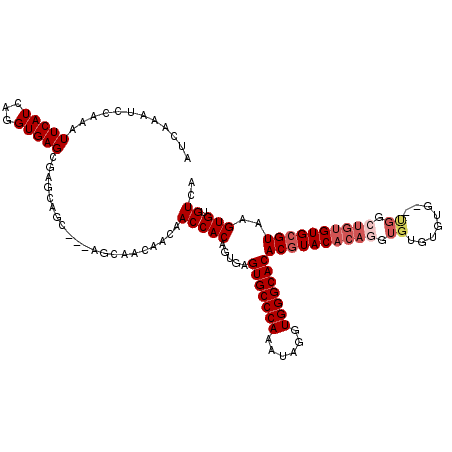
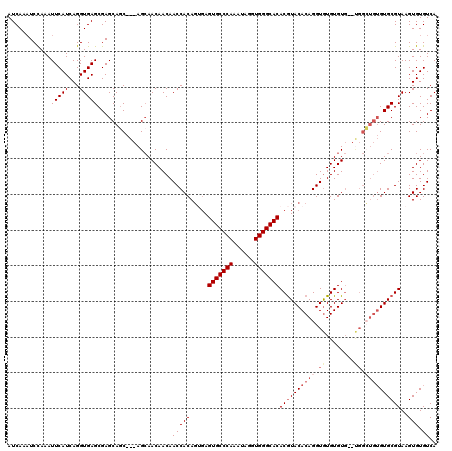
| Location | 2,993,397 – 2,993,512 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.64 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -30.64 |
| Energy contribution | -32.33 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.973052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2993397 115 + 22407834 ACAACAACCACAGUGAGUGCCCAAAUAGGUGGGCACACGUACACAGGUGUGUGUG--UGGCUGUGUGCGUAAGUGUGUCACUUGUUGUUCUUGGG---AACAUUAGAGAAAAGUAAAUAG ........((((.(..(((((((......)))))))((((((((((.(.......--.).)))))))))).).))))...(((..(((((....)---))))...)))............ ( -39.10) >DroSec_CAF1 5163 107 + 1 ACAACAACCACAGUGAGUGCCCAAAUAGGUGGGCACACGUACACAGGUAUGUGU----------GUGCGUAAGUGUGUCACUUGUUGUUCUUGGG---AACAUUAGAAAAAAGUAAAUAG .(((((.....((((((((((((......)))))))(((((((((......)))----------)))))).......))))))))))((((((..---..))..))))............ ( -35.80) >DroSim_CAF1 5137 115 + 1 ACAACAACCACAGAGAGUGCCCAAAUAGGUGGGCACACGUACACAGGUGUGUGUG--UGGCUGUGUGCGUAAGUGUGUCACUUGUUGUUCUUGGG---AACAUUAGAAAAAAGUAAAUAG .((((((.((((....(((((((......)))))))((((((((((.(.......--.).))))))))))...))))....))))))((((((..---..))..))))............ ( -38.70) >DroYak_CAF1 5168 118 + 1 ACAACAACCACAC--AGUGCCCAAAUAGGUGGGCACACAUACACAGGUGUGUGUGAGCGGGUGUGUGCGUAAGUGUGUCACUUGUUGUUCUUGGGCCGAGCAUUAGAAAAAAGUAAAUAG .((((.(((((((--((((((((......))))))).(((((((....)))))))......)))))).))(((((...)))))))))((((...((...))...))))............ ( -38.90) >consensus ACAACAACCACAGUGAGUGCCCAAAUAGGUGGGCACACGUACACAGGUGUGUGUG__UGGCUGUGUGCGUAAGUGUGUCACUUGUUGUUCUUGGG___AACAUUAGAAAAAAGUAAAUAG .((((((.((((....(((((((......)))))))((((((((((.((........)).))))))))))...))))....))))))((((..(......)...))))............ (-30.64 = -32.33 + 1.69)

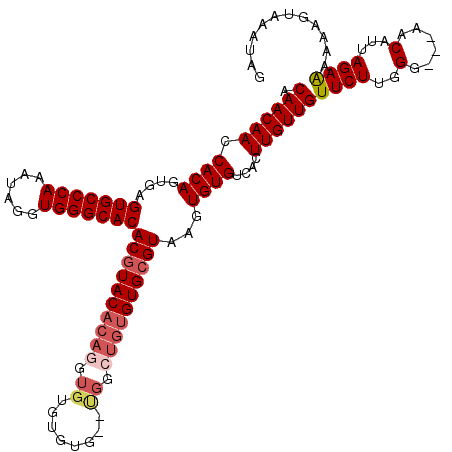
| Location | 2,993,397 – 2,993,512 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.64 |
| Mean single sequence MFE | -34.35 |
| Consensus MFE | -27.48 |
| Energy contribution | -27.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.26 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
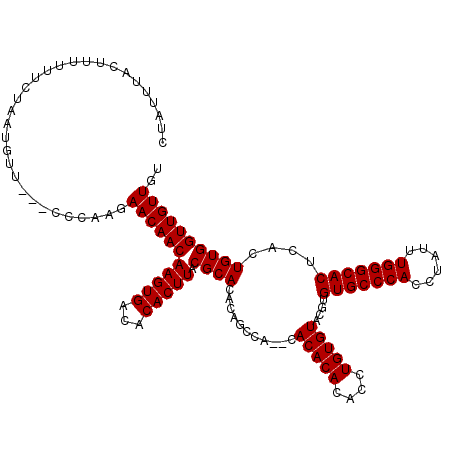
>2L_DroMel_CAF1 2993397 115 - 22407834 CUAUUUACUUUUCUCUAAUGUU---CCCAAGAACAACAAGUGACACACUUACGCACACAGCCA--CACACACACCUGUGUACGUGUGCCCACCUAUUUGGGCACUCACUGUGGUUGUUGU ..................((((---(....)))))..(((((...)))))..(((.(((((((--((.((((....))))..(.(((((((......))))))).)..)))))))))))) ( -36.50) >DroSec_CAF1 5163 107 - 1 CUAUUUACUUUUUUCUAAUGUU---CCCAAGAACAACAAGUGACACACUUACGCAC----------ACACAUACCUGUGUACGUGUGCCCACCUAUUUGGGCACUCACUGUGGUUGUUGU ......................---......((((((.(((((.......(((.((----------(((......))))).)))(((((((......))))))))))))...)))))).. ( -31.70) >DroSim_CAF1 5137 115 - 1 CUAUUUACUUUUUUCUAAUGUU---CCCAAGAACAACAAGUGACACACUUACGCACACAGCCA--CACACACACCUGUGUACGUGUGCCCACCUAUUUGGGCACUCUCUGUGGUUGUUGU ..................((((---(....)))))..(((((...)))))..(((.(((((((--((.((((....))))..(.(((((((......))))))).)..)))))))))))) ( -36.00) >DroYak_CAF1 5168 118 - 1 CUAUUUACUUUUUUCUAAUGCUCGGCCCAAGAACAACAAGUGACACACUUACGCACACACCCGCUCACACACACCUGUGUAUGUGUGCCCACCUAUUUGGGCACU--GUGUGGUUGUUGU ...............................(((((((((((...)))))...((((((...((..(((((....)))))..))(((((((......))))))))--))))))))))).. ( -33.20) >consensus CUAUUUACUUUUUUCUAAUGUU___CCCAAGAACAACAAGUGACACACUUACGCACACAGCCA__CACACACACCUGUGUACGUGUGCCCACCUAUUUGGGCACUCACUGUGGUUGUUGU ...............................(((((((((((...))))).((((...........(((((....)))))....(((((((......)))))))....)))))))))).. (-27.48 = -27.47 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:11 2006