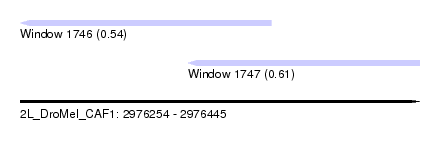
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,976,254 – 2,976,445 |
| Length | 191 |
| Max. P | 0.605837 |

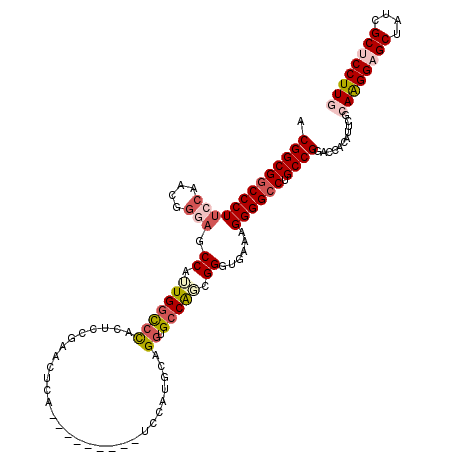
| Location | 2,976,254 – 2,976,374 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.75 |
| Mean single sequence MFE | -45.72 |
| Consensus MFE | -35.70 |
| Energy contribution | -37.06 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2976254 120 - 22407834 GGACUGCCGGAACACAUUCGCAAGGAGCUAUCGCUCCUUGGCCCCGCCAAGCGCAAGUGGUUCUUUCGCUAUCUGGACACGGGCAUCAGCAAUGAAAAGGCCCUUAUGAAGGCCUCGGAA ......(((.....(((((.((((((((....))))))))((((..(((((((.(((.....))).))))...)))....))))....).))))...(((((........)))))))).. ( -43.10) >DroSec_CAF1 6236 120 - 1 GGCCUGCCGGACCAUAUUCGCAGGGAGCUAACGCUCCUUGGCGCCACCAAGCGCAGGAGUUUCUUUCGCUAUCUGGACACGGGCAUCAGCAUUGAAAGGGCCCUCAUUAAGGCCUCGGAA (((((.(((((((.......((((((((....))))))))((((......)))).))(((.......))).)))))....((((.((......))....))))......)))))...... ( -46.20) >DroSim_CAF1 6231 120 - 1 GGCCUGCCGGACCAUAUUCGCAGGGAGCUAACGCUCCUUGGCGCCACCAAGCGCAGGAGUUUCUUUCGCUAUCUGGACACGGGCAUCAGCAUUGAAAGGGCCCUCAUUAAGGCCUCGGAA (((((.(((((((.......((((((((....))))))))((((......)))).))(((.......))).)))))....((((.((......))....))))......)))))...... ( -46.20) >DroEre_CAF1 6376 120 - 1 GGCCUGCCGGACCACAUUCUCAAGGAGCUUUCGCACCUUAGCGCCGCCAAGCGCAAGUGGUUCUUUCACUAUGUGGACAUGGGCUUCAGCAAUGAGAAGGCCCUCGCCAGGGCCUCGGAA (((((...(((((((......((((.((....)).)))).((((......))))..)))))))..........(((....((((((((....)))...)))))...))))))))...... ( -47.30) >DroYak_CAF1 6369 120 - 1 GGCCUGCCGGAACACAUACGCAAGGAGCUUUCGCACCUUAGCGCCGCCAAGCGCAAGUGGUUCUAUCGCUUCCUGGACGGGGGCUGCAGCAAUGAGAAGGCCCUCACCAAGGCCUUGGAA (((((((.(((((.(((....((((.((....)).)))).((((......))))..))))))))...))....(((..(((((((.((....))....))))))).))))))))...... ( -45.80) >consensus GGCCUGCCGGACCACAUUCGCAAGGAGCUAUCGCUCCUUGGCGCCGCCAAGCGCAAGUGGUUCUUUCGCUAUCUGGACACGGGCAUCAGCAAUGAAAAGGCCCUCAUCAAGGCCUCGGAA (((((.(((((.......((((((((((....))))))).)))......((((.(((.....))).)))).)))))....((((.(((....)))....))))......)))))...... (-35.70 = -37.06 + 1.36)

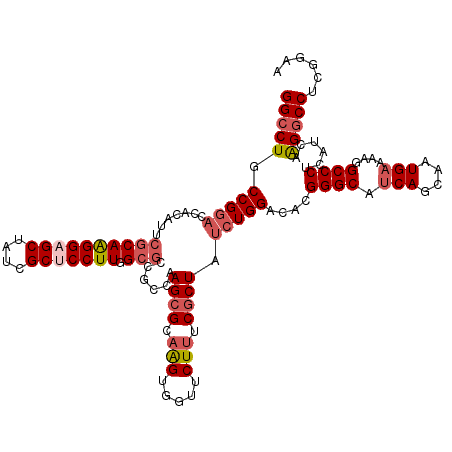
| Location | 2,976,334 – 2,976,445 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -46.39 |
| Consensus MFE | -32.83 |
| Energy contribution | -34.47 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2976334 111 - 22407834 ACGGCGGCCCUUCCGACGGGAGCCAUUGGCCCACUCCGAACUCA---------UCCAUGCAGGUGACAUCGGGUGAAAGGGGACUGCCGGAACACAUUCGCAAGGAGCUAUCGCUCCUUG .(((((((((((((....)))(((...))).(((.((((..(((---------((......)))))..)))))))...)))).))))))...........((((((((....)))))))) ( -47.30) >DroSec_CAF1 6316 111 - 1 ACGGCGGCCCUUCCAACGGGAGCCAUUGGCCCACUUCGAACUCA---------UCCAUGCAGGUGCCAGCGGGUGAAAGGGGCCUGCCGGACCAUAUUCGCAGGGAGCUAACGCUCCUUG .((((((((((......))).)))...(((((.(((.....(((---------(((..((.(....).)))))))))))))))).))))...........((((((((....)))))))) ( -49.80) >DroSim_CAF1 6311 111 - 1 ACGGCGGCCCUUCCAACGGGAGCCAUUGGCCCACUUCGAACUCA---------UCCAUGCAGGUGCCAGCGGGUAAAGGGGGCCUGCCGGACCAUAUUCGCAGGGAGCUAACGCUCCUUG .(((((((((((((....)))(((.(((((((............---------........)).)))))..)))....)))))).))))...........((((((((....)))))))) ( -46.65) >DroEre_CAF1 6456 120 - 1 ACGGCGGCCCUGCCAAUGCCAGCCAGUGGUCUAAUCCGAACCCAAAAAAGCAGUCCAUGCCGUUGCCACCGGAGGAUAGGGGCCUGCCGGACCACAUUCUCAAGGAGCUUUCGCACCUUA .((((((((((.((...((((.....))))....((((....(((....(((.....)))..)))....))))))...)))))).))))............((((.((....)).)))). ( -40.00) >DroYak_CAF1 6449 120 - 1 ACGGCGGCCCUUCCAACGGCAGCCAGUGGUCCACUGCAAGCCCAAACGUGCAGUCCGUGCAGGUGCCGGCGGAGGAGAGGGGCCUGCCGGAACACAUACGCAAGGAGCUUUCGCACCUUA .(((((((((((((..(((((.((.((((...((((((.(......).))))))))..)).)))))))..))).....)))))).))))............((((.((....)).)))). ( -48.20) >consensus ACGGCGGCCCUUCCAACGGGAGCCAUUGGCCCACUCCGAACUCA_________UCCAUGCAGGUGCCAGCGGGUGAAAGGGGCCUGCCGGACCACAUUCGCAAGGAGCUAUCGCUCCUUG .(((((((((((((....))).((.(((((((.............................)).))))).))......)))))).))))...........((((((((....)))))))) (-32.83 = -34.47 + 1.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:58 2006