| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,975,860 – 2,976,084 |
| Length | 224 |
| Max. P | 0.787265 |

| Location | 2,975,860 – 2,975,973 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -30.09 |
| Consensus MFE | -24.64 |
| Energy contribution | -25.36 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2975860 113 + 22407834 CCAGCCAUUGGCGGGAGUUCUUGUCCAUGCAGACAACUUGGAUCAUUCCAGUAAGAAAUUGAAUGCCCCCAAAAUUGAUGCUGGACUUCCAACCCUUUUUCAACUCCUCCACA ........(((.(((((((.(((((......))))).(((((....(((((((...((((............))))..)))))))..))))).........)))))))))).. ( -28.40) >DroSec_CAF1 5842 113 + 1 CCAGCCACUCGCGGGAGUUCUUGUCCAUGCAGACAACUUGGAUCAUUCCAGUAAGGAAUUCGAUGCCCCAAAAGUUGAUGCUGGACUUCCAACCCUUUUUCAACUCCUCCACA ..........(.(((((((...(((((.(((..((((((((((((((((.....)))))..))).))....)))))).))))))))...............))))))).)... ( -29.57) >DroSim_CAF1 5837 113 + 1 CCAGCCACUCGCGGGAGUUCUUGUCCAUGCAGACAACUUGGAUCAUUCCAGUAAGGAAUUCGAUGCCCCAAAAGUUGAUGCUGGACUUCCAACCCUUUUUCAACUCCUCCACA ..........(.(((((((...(((((.(((..((((((((((((((((.....)))))..))).))....)))))).))))))))...............))))))).)... ( -29.57) >DroEre_CAF1 5979 113 + 1 CCAGCCAUUGGCGGGAGUUCUUGUCCAUGCAGACGAUUUGGAUCAGUCCGGAAAGGAAUUGGAUGCCCCCAAAGUUGAUGCUGGACUUCCAACCCUUUUUCAUUUCCUCCACA ........(((.((((((..(((((......))))).(((((..(((((((...((..((((......))))..))....)))))))))))).........)))))).))).. ( -30.90) >DroYak_CAF1 5942 113 + 1 CCAGCCAUUGGCGGGAGUUCUUGUCCAUGCAGACAACUUGGAUCAGUCCAGAGAGGAAUUGGAUGCCCCCAAAGUUGAUGCUGGACUUCCAACCCUUUUUCAUUUCCUCCACC ...(((...)))(((.......(((((.(((..((((((((....((((((.......))))))....)).)))))).))))))))......))).................. ( -32.02) >consensus CCAGCCAUUGGCGGGAGUUCUUGUCCAUGCAGACAACUUGGAUCAUUCCAGUAAGGAAUUGGAUGCCCCCAAAGUUGAUGCUGGACUUCCAACCCUUUUUCAACUCCUCCACA ...((.....))(((.......(((((.(((..((((((((((((((((.....)))))..))).))....)))))).))))))))......))).................. (-24.64 = -25.36 + 0.72)


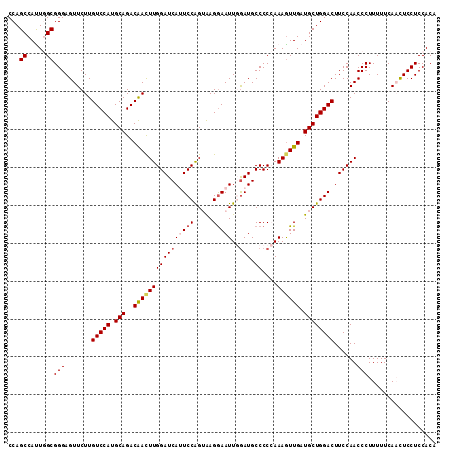
| Location | 2,975,973 – 2,976,084 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -35.26 |
| Consensus MFE | -20.96 |
| Energy contribution | -20.64 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.723080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2975973 111 + 22407834 AGGCCGGCUUCGAUCUCAUGCAAUUGCUGAUUAGUCAGCUCGCAAUCCGGAUAAGUUUCCGCCAUUAUUGCCAUGGUAAUACUAUUCUGUUUCGAGG------UCCAU---GACGGCUCU .(((((.....((((((.(((....(((((....)))))..)))...((((......))))....((((((....))))))............))))------))...---..))))).. ( -34.10) >DroSec_CAF1 5955 111 + 1 AGGCCGGCUUCGAUCUCUUGCAAUUGCUGAUUGGUCAGCUCGCAAUCCGGAUAAUUUUCCGCCAUUACUGUCAUGGUAAUACUAUUCUGAUUGAAGG------CCCAU---AACGGCUCU .((((((((((((((..((((....(((((....)))))..))))..((((......))))..((((((.....))))))........))))))).)------))...---...)))).. ( -32.00) >DroSim_CAF1 5950 111 + 1 AGGCCGGCUUCGAUCUCUUGCAAUUGCUGAUUGGUCAGCUCGCAAUCCGGAUAAUUUUCCGCCAUUACUGCCAUGGUAAUACUAUUCUGAUUGAAGG------UCCAU---AACGGCCCU .(((((.((((((((..((((....(((((....)))))..))))..((((......))))..((((((.....))))))........)))))))).------.....---..))))).. ( -33.90) >DroEre_CAF1 6092 120 + 1 AGACCGUCUUCGAUCUCAUGCAGUUGCUUAUUAGUGAGCACGCAAUCGGGAUAAUUUUCCGCCAUUACUGCCAUGGUAAAACCAUUUUGCUUGGAGCCCAGUGCCCUUGAAGACGGGUCC ...((((((((((...((((((((((((((....))))))......(((((.....))))).....)))).))))(((((.....)))))..((.((.....)))))))))))))).... ( -36.70) >DroYak_CAF1 6055 120 + 1 AAGCCGUCUUCGAUCUCAUGCAGUUGCUUAUUAGUCAGCAAGCAAUCCGGAUAAGUUUCUGCCAUAAUUGCCAUGGUAAGACCAUUCUGCUUCGUGCUUAGUGCCCGUGAAGACGGCUCU .(((((((((((....(((...((((((....)).))))(((((...((((...((((.((((((.......))))))))))...)))).....))))).)))....))))))))))).. ( -39.60) >consensus AGGCCGGCUUCGAUCUCAUGCAAUUGCUGAUUAGUCAGCUCGCAAUCCGGAUAAUUUUCCGCCAUUACUGCCAUGGUAAUACUAUUCUGAUUGGAGG______CCCAU___GACGGCUCU .(((((............(((....(((((....)))))..)))....(((......)))(((((.......)))))....................................))))).. (-20.96 = -20.64 + -0.32)

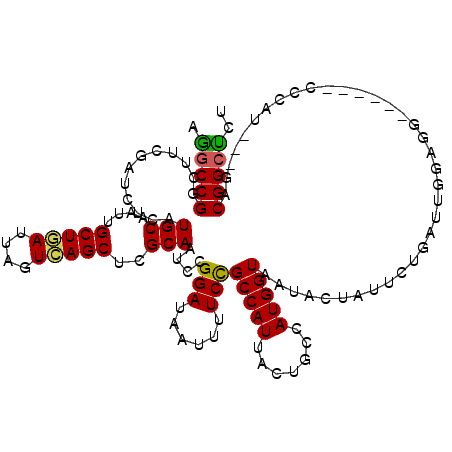
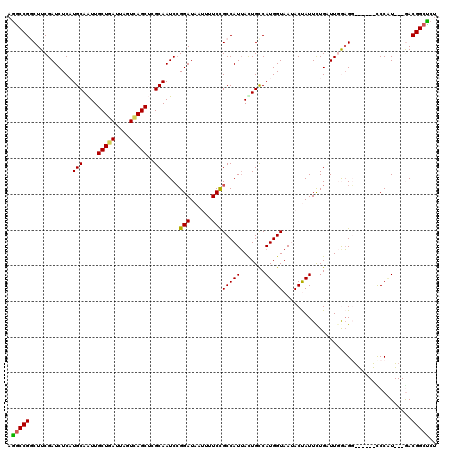
| Location | 2,975,973 – 2,976,084 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -36.81 |
| Consensus MFE | -16.14 |
| Energy contribution | -17.74 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2975973 111 - 22407834 AGAGCCGUC---AUGGA------CCUCGAAACAGAAUAGUAUUACCAUGGCAAUAAUGGCGGAAACUUAUCCGGAUUGCGAGCUGACUAAUCAGCAAUUGCAUGAGAUCGAAGCCGGCCU ((.((((((---(((((------(.((......))...))....)))))))......((((((......))).(((((((((((((....)))))..)))))....)))...)))))))) ( -32.00) >DroSec_CAF1 5955 111 - 1 AGAGCCGUU---AUGGG------CCUUCAAUCAGAAUAGUAUUACCAUGACAGUAAUGGCGGAAAAUUAUCCGGAUUGCGAGCUGACCAAUCAGCAAUUGCAAGAGAUCGAAGCCGGCCU ((.((((((---((((.------.....................)))))))......((((((......)))...(((((((((((....)))))..)))))).........)))))))) ( -32.45) >DroSim_CAF1 5950 111 - 1 AGGGCCGUU---AUGGA------CCUUCAAUCAGAAUAGUAUUACCAUGGCAGUAAUGGCGGAAAAUUAUCCGGAUUGCGAGCUGACCAAUCAGCAAUUGCAAGAGAUCGAAGCCGGCCU .((((((((---(((((------(.(((.....)))..))....)))))))......((((((......)))...(((((((((((....)))))..)))))).........)))))))) ( -35.00) >DroEre_CAF1 6092 120 - 1 GGACCCGUCUUCAAGGGCACUGGGCUCCAAGCAAAAUGGUUUUACCAUGGCAGUAAUGGCGGAAAAUUAUCCCGAUUGCGUGCUCACUAAUAAGCAACUGCAUGAGAUCGAAGACGGUCU (((((.((((((..((((((..........((...((((.....)))).)).(((((.(.(((......)))).)))))))))))........((....))........))))))))))) ( -42.50) >DroYak_CAF1 6055 120 - 1 AGAGCCGUCUUCACGGGCACUAAGCACGAAGCAGAAUGGUCUUACCAUGGCAAUUAUGGCAGAAACUUAUCCGGAUUGCUUGCUGACUAAUAAGCAACUGCAUGAGAUCGAAGACGGCUU .(((((((((((.((.((.....)).))..((((.((((.....))))(((((((.(((.(........)))))))))))((((........)))).))))........))))))))))) ( -42.10) >consensus AGAGCCGUC___AUGGG______CCUCCAAGCAGAAUAGUAUUACCAUGGCAGUAAUGGCGGAAAAUUAUCCGGAUUGCGAGCUGACUAAUCAGCAAUUGCAUGAGAUCGAAGCCGGCCU ...((((.....................................((((.......))))((((......))))(((((((((((((....)))))..)))))....))).....)))).. (-16.14 = -17.74 + 1.60)

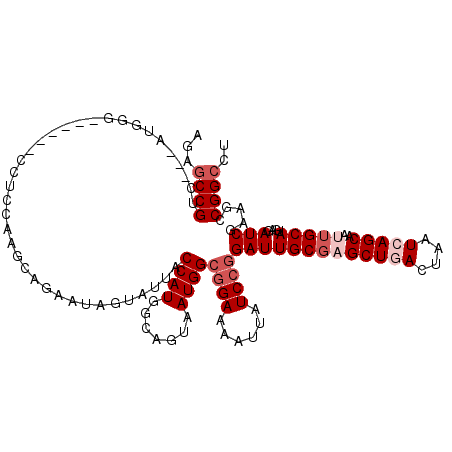
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:55 2006