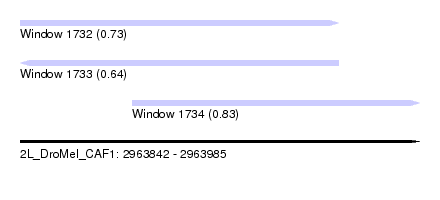
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,963,842 – 2,963,985 |
| Length | 143 |
| Max. P | 0.827928 |

| Location | 2,963,842 – 2,963,956 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -21.25 |
| Energy contribution | -21.03 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.727294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2963842 114 + 22407834 ACGAAAUUUAUUUACAACAAGCCCCAGAACCAAUACGAGUUGUUGCAGCAGCAGCUGCAGCUGCAGCAGGAGUUCGCCCAGUUUUUGACCAAUGUGGCCAACAGCG------CCGCAACG ........................(((((.(....((((((((((((((.((....)).))))))))))...))))....).))))).....(((((((....).)------)))))... ( -30.20) >DroVir_CAF1 22624 114 + 1 ACGAAAUUCAUAUACAAUAAGCCACAAAAUCAUUUCGAGCUACUGCAACAGCAGCUGCAGCUGCAGCAGGAAUUUGCCCAGUUCCUCACAAAUGUGGCAAACAGUG------CAGCUGGG ...((((((..........((((.............).))).((((....(((((....))))).)))))))))).(((((((...(((....)))(((.....))------)))))))) ( -30.52) >DroGri_CAF1 20215 114 + 1 ACGAAAUUCAUUUACAAUAAGCCACAAAAUCAUUUCGAGCUGCUGCAACAGCAGCUGCAACUGCAGCAGGAAUUUGCACAGUUCCUUACGAAUGUGGCCAAUAGUG------CAACUGGG ............(((.((..((((((...((.....((((((.(((((...(.(((((....)))))..)...))))))))))).....)).))))))..)).)))------........ ( -31.00) >DroMoj_CAF1 23091 114 + 1 ACGAAAUUCAUCUACAAUAAGCCGCAGAAUCAUUUCGAAUUGCUACAACAGCAGCUGCAGCUGCAGCAGGAGUUUGCUCAGUUCCUUACCAAUGUGGCUAAUAGUG------CAGCUGGA ......((((.((.((...((((((((((....)))((((((........(((((....)))))(((((....)))))))))))........))))))).....))------.)).)))) ( -30.50) >DroAna_CAF1 17169 120 + 1 ACGAAAUUUGUAUACAACAAGCCCCAAAAUCACUUUGAGUUGUUGCAGCAGCAACUGCAGCUGCAGCAGGAGUUCGCCCAGUUCUUAACUAACGUGGCCAACAGCGCUGCUGCCGCAGCU ......(((((.....)))))...((((.....))))((((((.((((((((..((((.......))))..(((.(((..(((.......)))..))).)))...)))))))).)))))) ( -37.30) >DroPer_CAF1 124948 114 + 1 ACAAAAUUCAUUUACAACAAGCCACAAAAUCAAUUUGAGUUGCUCCAGCAACAGCUGCAACUGCAGCAGGAAUUCGCCCAGUUCCUGACAAACGUGGCCAGCAGUG------CAGUAGGA ..........(((((.....(((((.............(((((....))))).(((((....)))))(((((((.....))))))).......)))))..((...)------).))))). ( -32.50) >consensus ACGAAAUUCAUUUACAACAAGCCACAAAAUCAUUUCGAGUUGCUGCAACAGCAGCUGCAGCUGCAGCAGGAAUUCGCCCAGUUCCUGACAAAUGUGGCCAACAGUG______CAGCAGGG ....................(((((.............((((......)))).(((((....)))))(((((((.....))))))).......)))))...................... (-21.25 = -21.03 + -0.22)

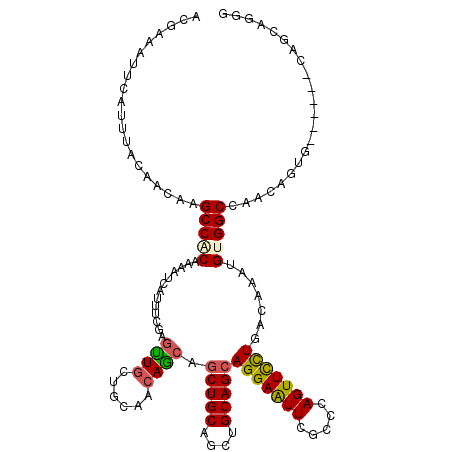
| Location | 2,963,842 – 2,963,956 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -22.09 |
| Energy contribution | -20.85 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2963842 114 - 22407834 CGUUGCGG------CGCUGUUGGCCACAUUGGUCAAAAACUGGGCGAACUCCUGCUGCAGCUGCAGCUGCUGCUGCAACAACUCGUAUUGGUUCUGGGGCUUGUUGUAAAUAAAUUUCGU .(((((((------((..(((.(((.((.(........).))))).)))...)))))))))((((((....))))))(((((.......((((....)))).)))))............. ( -37.00) >DroVir_CAF1 22624 114 - 1 CCCAGCUG------CACUGUUUGCCACAUUUGUGAGGAACUGGGCAAAUUCCUGCUGCAGCUGCAGCUGCUGUUGCAGUAGCUCGAAAUGAUUUUGUGGCUUAUUGUAUAUGAAUUUCGU ..((((((------(((.(((((((.((.((......)).)))))))))....).)))))))).((((((((...))))))))((((((.....((..(....)..)).....)))))). ( -35.70) >DroGri_CAF1 20215 114 - 1 CCCAGUUG------CACUAUUGGCCACAUUCGUAAGGAACUGUGCAAAUUCCUGCUGCAGUUGCAGCUGCUGUUGCAGCAGCUCGAAAUGAUUUUGUGGCUUAUUGUAAAUGAAUUUCGU ..((.(((------((.....(((((((.((((..(.(((((((((......))).)))))).)((((((((...))))))))....))))...)))))))...))))).))........ ( -40.30) >DroMoj_CAF1 23091 114 - 1 UCCAGCUG------CACUAUUAGCCACAUUGGUAAGGAACUGAGCAAACUCCUGCUGCAGCUGCAGCUGCUGUUGUAGCAAUUCGAAAUGAUUCUGCGGCUUAUUGUAGAUGAAUUUCGU ..((((((------(.......(((.....))).........((((......)))))))))))..(((((....)))))....((((((...(((((((....)))))))...)))))). ( -37.30) >DroAna_CAF1 17169 120 - 1 AGCUGCGGCAGCAGCGCUGUUGGCCACGUUAGUUAAGAACUGGGCGAACUCCUGCUGCAGCUGCAGUUGCUGCUGCAACAACUCAAAGUGAUUUUGGGGCUUGUUGUAUACAAAUUUCGU (((((((((.((((((..(((.(((..(((.......)))..))).)))...)))))).))))))))).....(((((((.(((((((...)))))))...)))))))............ ( -44.70) >DroPer_CAF1 124948 114 - 1 UCCUACUG------CACUGCUGGCCACGUUUGUCAGGAACUGGGCGAAUUCCUGCUGCAGUUGCAGCUGUUGCUGGAGCAACUCAAAUUGAUUUUGUGGCUUGUUGUAAAUGAAUUUUGU ......((------((..((.(((((((...(((((....((((....((((.((.(((((....))))).)).))))...))))..)))))..))))))).))))))............ ( -33.70) >consensus CCCAGCUG______CACUGUUGGCCACAUUGGUAAGGAACUGGGCAAACUCCUGCUGCAGCUGCAGCUGCUGCUGCAGCAACUCGAAAUGAUUUUGUGGCUUAUUGUAAAUGAAUUUCGU ...............((.((.((((.((...(((........))).......(((((((((.((....)).)))))))))..............)).)))).)).))............. (-22.09 = -20.85 + -1.24)
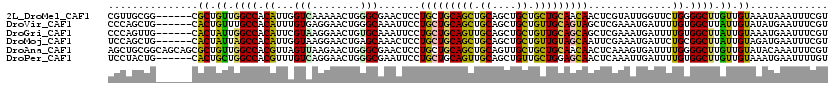
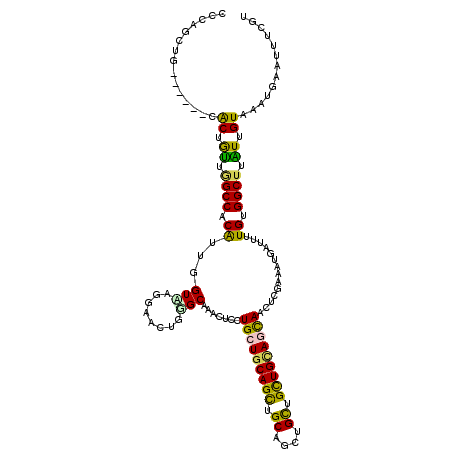
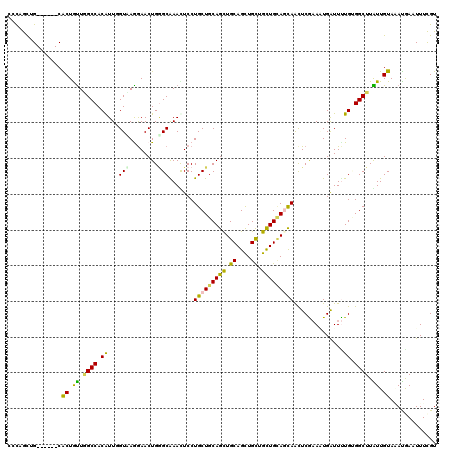
| Location | 2,963,882 – 2,963,985 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.61 |
| Mean single sequence MFE | -36.55 |
| Consensus MFE | -19.25 |
| Energy contribution | -19.17 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
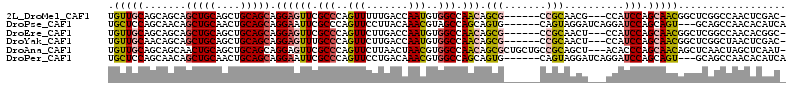
>2L_DroMel_CAF1 2963882 103 + 22407834 UGUUGCAGCAGCAGCUGCAGCUGCAGCAGGAGUUCGCCCAGUUUUUGACCAAUGUGGCCAACAGCG------CCGCAACG---CCAUCCAGCAACGGCUCGGCCAACUCGAC- (((((((((.((....)).))))))))).(((((.(((.(((..((......(((((((....).)------)))))..(---(......))))..))).))).)))))...- ( -41.30) >DroPse_CAF1 123286 104 + 1 UGCUCCAGCAACAGCUGCAACUGCAGCAGGAAUUCGCCCAGUUCCUUACAAACGUAGCCAGCAGUG------CAGUAGGAUCAGGAUCCAGCAGU---GCAGCCAACACAUCA (((....)))...((((((.((((.(((((((((.....))))))((((....)))).......))------)....((((....)))).)))))---))))).......... ( -31.50) >DroEre_CAF1 16395 103 + 1 UGUUGCAGCAGCAGCUGCAGCUGCAGCAGGAGUUCGCCCAGUUCUUGACCAAUGUGGCCAACAGCG------CCGCAACU---CCAUCCAGCAACGGCUCGGCCAACACGGC- .(((((.((.(((((....))))).)).((((((.((...(((.(((.((.....)).))).))).------..))))))---)).....)))))(((...)))........- ( -39.00) >DroYak_CAF1 16577 103 + 1 UGUUGCAACAGCAGCUGCAGCUGCAGCAGGAGUUUGCCCAGUUCUUGACCAAUGUGGCCAACAGCG------CCGCAACU---CCAUCCAGCAACGGCUCGGCUAACUCGAC- ...(((....)))(((((....)))))..(((((.(((.(((..(((.....(((((((....).)------))))).((---......)))))..))).))).)))))...- ( -34.80) >DroAna_CAF1 17209 109 + 1 UGUUGCAGCAGCAACUGCAGCUGCAGCAGGAGUUCGCCCAGUUCUUAACUAACGUGGCCAACAGCGCUGCUGCCGCAGCU---ACACCCAGCAACAGCUCAACUAGCUCAAU- ((((((.((((...))))((((((.((((.((..(((...(((....((....))....))).))))).)))).))))))---.......))))))(((.....))).....- ( -38.10) >DroPer_CAF1 124988 104 + 1 UGCUCCAGCAACAGCUGCAACUGCAGCAGGAAUUCGCCCAGUUCCUGACAAACGUGGCCAGCAGUG------CAGUAGGAUCAGGAUCCAGCAGU---GCAGCCAACACAUCA (((....)))...(((((....)))))(((((((.....)))))))((......((((..(((.((------(....((((....)))).))).)---)).)))).....)). ( -34.60) >consensus UGUUGCAGCAGCAGCUGCAGCUGCAGCAGGAGUUCGCCCAGUUCUUGACCAACGUGGCCAACAGCG______CCGCAACU___CCAUCCAGCAACGGCUCAGCCAACACAAC_ .(((((.......(((((....))))).((((((.(((..(((.......)))..))).))).(((.......)))..........))).))))).................. (-19.25 = -19.17 + -0.08)
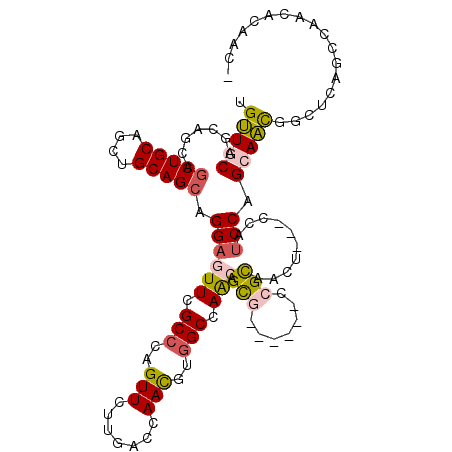
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:47 2006