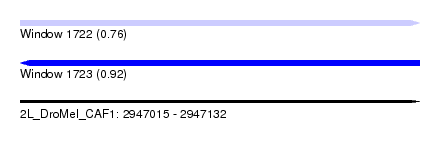
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,947,015 – 2,947,132 |
| Length | 117 |
| Max. P | 0.921003 |

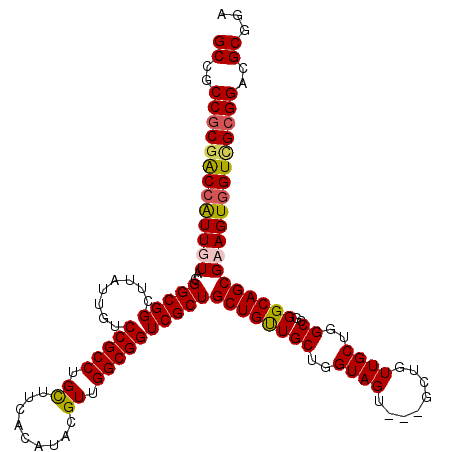
| Location | 2,947,015 – 2,947,132 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.98 |
| Mean single sequence MFE | -46.80 |
| Consensus MFE | -36.61 |
| Energy contribution | -36.67 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2947015 117 + 22407834 GCCGCGUCCGCGACCACUUCGCUGCCCGGCCAGCAACAGC---ACUACCAGCAACAGCAGCGACCGCCUACGUAUGUGAAGCAGGCGGACAAUAAGCCGCCGUACAAUGGUCGCGGCGGC (((((...(((((((((((((((((..(((..((....))---.......((....)).......)))...))).))))))..(((((........)))))......))))))))))))) ( -48.50) >DroSim_CAF1 63210 117 + 1 GCCGCGUCCGCGACCACUUCGCUGCCCGGCCAGCAACAGC---ACUACCAGCAACAGCAGCGACCGCCAACGUAUGUGAAGCAGGCGGACAAUAAGCCGCCGUACAAUGGUCGCGGCGGC (((((...(((((((((((((((((..(((..((....))---.......((....)).......)))...))).))))))..(((((........)))))......))))))))))))) ( -48.50) >DroEre_CAF1 64280 120 + 1 UCCGCAUCCGCCACCACUGCGCUGCCCGGCCAGCAACAACAGCACUACCAGCAGCAGCAGCGACCGCCGACGUAUGUGAAACAGGCGGACAAUAAGCCGCCGUACAACGGCCGUGGCGGC .......((((((((.(((((((((..((...((.......))....)).))))).)))).)...((((..(((((.....).(((((........)))))))))..)))).))))))). ( -49.00) >DroYak_CAF1 64373 117 + 1 UCCGCAUCCGCAACGACUGCGCUGCCCGGCCAGCAACAAC---ACUACCAGCAACAGCAGCGACCGCCAACGUAUGUGAAGCAGGCGGAUAAUAAGCCGCCGUACAACGGUCGCGGCGGC .....((((((.....((((((((......)))).....(---(((((..((....)).((....))....))).)))..)))))))))).....((((((((((....)).)))))))) ( -41.20) >consensus GCCGCAUCCGCGACCACUGCGCUGCCCGGCCAGCAACAAC___ACUACCAGCAACAGCAGCGACCGCCAACGUAUGUGAAGCAGGCGGACAAUAAGCCGCCGUACAACGGUCGCGGCGGC ...((....)).........(((((((((((.((.(((............((....)).(((........))).)))...)).(((((........))))).......))))).)))))) (-36.61 = -36.67 + 0.06)

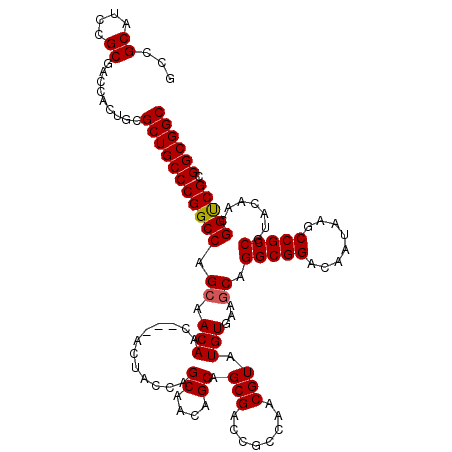
| Location | 2,947,015 – 2,947,132 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.98 |
| Mean single sequence MFE | -59.10 |
| Consensus MFE | -48.01 |
| Energy contribution | -48.33 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2947015 117 - 22407834 GCCGCCGCGACCAUUGUACGGCGGCUUAUUGUCCGCCUGCUUCACAUACGUAGGCGGUCGCUGCUGUUGCUGGUAGU---GCUGUUGCUGGCCGGGCAGCGAAGUGGUCGCGGACGCGGC (((((((((((((((....((((((.....(.((((((((.........)))))))).)))))))(((((((((...---((....))..))).))))))..))))))))))...))))) ( -62.40) >DroSim_CAF1 63210 117 - 1 GCCGCCGCGACCAUUGUACGGCGGCUUAUUGUCCGCCUGCUUCACAUACGUUGGCGGUCGCUGCUGUUGCUGGUAGU---GCUGUUGCUGGCCGGGCAGCGAAGUGGUCGCGGACGCGGC ((((((((((((((((((.((((((.....).))))))))........((....)).((((((((...((..((((.---....))))..))..))))))))))))))))))...))))) ( -59.10) >DroEre_CAF1 64280 120 - 1 GCCGCCACGGCCGUUGUACGGCGGCUUAUUGUCCGCCUGUUUCACAUACGUCGGCGGUCGCUGCUGCUGCUGGUAGUGCUGUUGUUGCUGGCCGGGCAGCGCAGUGGUGGCGGAUGCGGA .(((((((((((((((.((((((((.....).))))).((.......))))))))))))(((((.(((((((((...((.......))..))).))))))))))).)))))))....... ( -62.10) >DroYak_CAF1 64373 117 - 1 GCCGCCGCGACCGUUGUACGGCGGCUUAUUAUCCGCCUGCUUCACAUACGUUGGCGGUCGCUGCUGUUGCUGGUAGU---GUUGUUGCUGGCCGGGCAGCGCAGUCGUUGCGGAUGCGGA ....(((((.((((...(((((.((.......(((((.((.........)).)))))..((((((...((..((((.---....))))..))..)))))))).))))).)))).))))). ( -52.80) >consensus GCCGCCGCGACCAUUGUACGGCGGCUUAUUGUCCGCCUGCUUCACAUACGUUGGCGGUCGCUGCUGUUGCUGGUAGU___GCUGUUGCUGGCCGGGCAGCGAAGUGGUCGCGGACGCGGA ((..(((((((((((((..(((((........(((((.((.........)).))))))))))((((((((..((((........))))..))..)))))))))))))))))))..))... (-48.01 = -48.33 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:35 2006