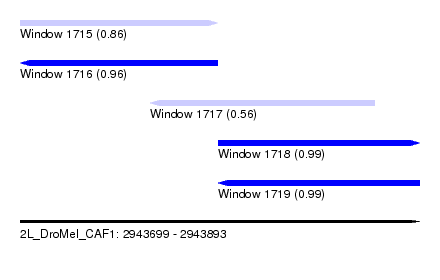
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,943,699 – 2,943,893 |
| Length | 194 |
| Max. P | 0.991798 |

| Location | 2,943,699 – 2,943,795 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 92.13 |
| Mean single sequence MFE | -22.46 |
| Consensus MFE | -17.83 |
| Energy contribution | -19.32 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2943699 96 + 22407834 AUUAAAUGUUUGUCUCUGUGGCUGCGCCGCCACUCCUUCAGUUGAGAUGUUAACAAAAUUUAUAAGACAAAUAAAUUGUAGCAGCCAGAGCACAAA------- ........(((((((((..((((((...............(((((....)))))..(((((((.......)))))))...)))))))))).)))))------- ( -22.50) >DroSim_CAF1 60570 96 + 1 AUUAAAUGUUUGUCUCUCUGGCUGCGCCGCCACUCCUUCAGUUGAGAUGUUAACAAAAUUUAUAAGACAAAUAAAUUGUAGCAGCCAGAGCACAAA------- ........(((((..((((((((((...............(((((....)))))..(((((((.......)))))))...)))))))))).)))))------- ( -26.30) >DroEre_CAF1 60956 103 + 1 AUUAAAUGCUUGUCUCCCUGGCUGCGCCGCCACUGCUUCAGUUGAGAUGUUAACAAAAUUUAUAAGACAAAUAAAUUGUAGCAGCAAGAACACAAAAAAAAAA ........((((......((((......))))(((((.(((((....((((..............))))....))))).)))))))))............... ( -18.64) >DroYak_CAF1 61038 96 + 1 AUUAAAUGUUUGUCUCUCUGGCUGCUGCGCCACUCCUUCAGUUGAGAUGUUAACAAAAUUUAUAAGACAAAUAAAUUGUAGCAGCAAGAACACAAA------- ........(((((...(((.((((((((............(((((....)))))..(((((((.......))))))))))))))).)))..)))))------- ( -22.40) >consensus AUUAAAUGUUUGUCUCUCUGGCUGCGCCGCCACUCCUUCAGUUGAGAUGUUAACAAAAUUUAUAAGACAAAUAAAUUGUAGCAGCAAGAACACAAA_______ .........((((..((((((((((...............(((((....)))))..(((((((.......)))))))...)))))))))).))))........ (-17.83 = -19.32 + 1.50)

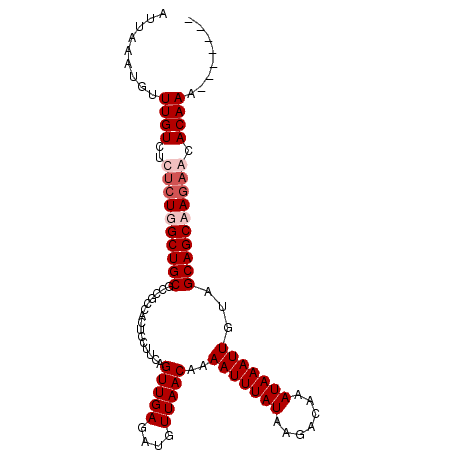
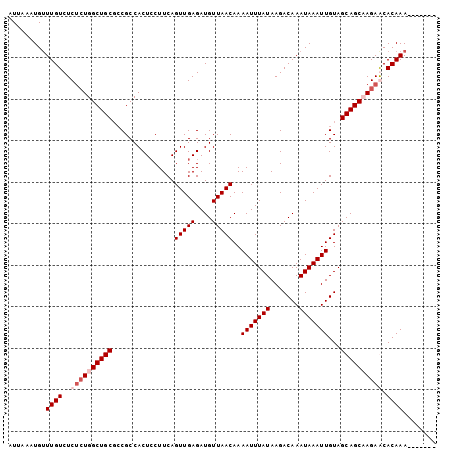
| Location | 2,943,699 – 2,943,795 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 92.13 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -22.45 |
| Energy contribution | -23.33 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2943699 96 - 22407834 -------UUUGUGCUCUGGCUGCUACAAUUUAUUUGUCUUAUAAAUUUUGUUAACAUCUCAACUGAAGGAGUGGCGGCGCAGCCACAGAGACAAACAUUUAAU -------(((((.((((((((((...(((((((.......)))))))..(((..((.(((........)))))..)))))))))..)))))))))........ ( -25.60) >DroSim_CAF1 60570 96 - 1 -------UUUGUGCUCUGGCUGCUACAAUUUAUUUGUCUUAUAAAUUUUGUUAACAUCUCAACUGAAGGAGUGGCGGCGCAGCCAGAGAGACAAACAUUUAAU -------(((((.((((((((((...(((((((.......)))))))..(((..((.(((........)))))..)))))))))))))..)))))........ ( -30.80) >DroEre_CAF1 60956 103 - 1 UUUUUUUUUUGUGUUCUUGCUGCUACAAUUUAUUUGUCUUAUAAAUUUUGUUAACAUCUCAACUGAAGCAGUGGCGGCGCAGCCAGGGAGACAAGCAUUUAAU .........(((((..(..(((((((((...((((((...)))))).)))).....((......)))))))..)..)))))((...(....)..))....... ( -22.70) >DroYak_CAF1 61038 96 - 1 -------UUUGUGUUCUUGCUGCUACAAUUUAUUUGUCUUAUAAAUUUUGUUAACAUCUCAACUGAAGGAGUGGCGCAGCAGCCAGAGAGACAAACAUUUAAU -------(((((.((((.((((((.((((((((.......)))))))..((((...(((........))).))))).)))))).))))..)))))........ ( -21.10) >consensus _______UUUGUGCUCUGGCUGCUACAAUUUAUUUGUCUUAUAAAUUUUGUUAACAUCUCAACUGAAGGAGUGGCGGCGCAGCCAGAGAGACAAACAUUUAAU .......(((((.((((((((((...(((((((.......)))))))..(((..((.(((........)))))..)))))))))))))..)))))........ (-22.45 = -23.33 + 0.87)

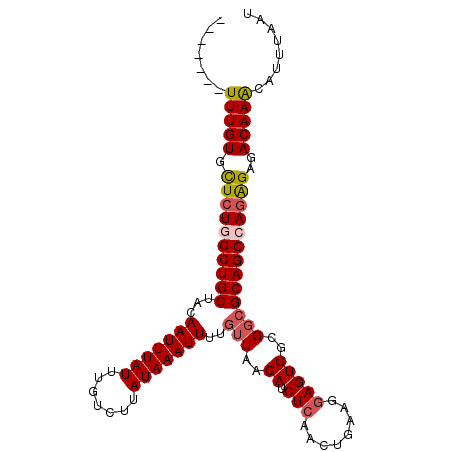
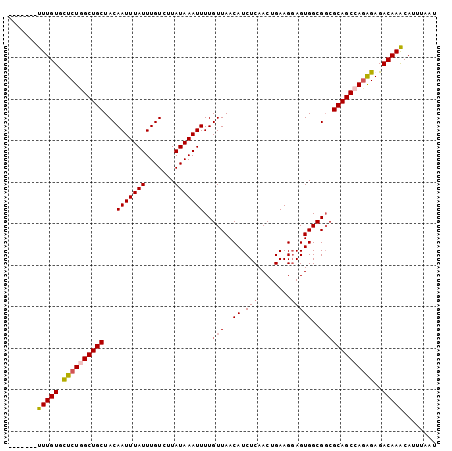
| Location | 2,943,762 – 2,943,871 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.78 |
| Mean single sequence MFE | -30.06 |
| Consensus MFE | -21.92 |
| Energy contribution | -21.66 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2943762 109 - 22407834 -CCCUUUUUCUUGGUCGGAAAAAAAAGGGACAGAAUGUGAGCAGUCACAAAUCUGCUGGCUUUUGGGCAUUCGGUUU----------UUUGUGCUCUGGCUGCUACAAUUUAUUUGUCUU -.(((((((.((......)).)))))))((((((...((((((((((((((.(((...(((....)))...)))...----------)))))).....)))))).)).....)))))).. ( -29.00) >DroSim_CAF1 60633 110 - 1 CCCCUUUUUCUCGGUCGGAAAAAAAGGGGACAGAAUGUGUGCAGUCACAAAUCUGCUGGCUUUUGGGCAUUCGGUUU----------UUUGUGCUCUGGCUGCUACAAUUUAUUUGUCUU .((((((((.((.....)).))))))))((((((...((((((((((((((.(((...(((....)))...)))...----------)))))).....))))).))).....)))))).. ( -34.30) >DroEre_CAF1 61019 117 - 1 --CUAUUUUCUCGGUCGGCAA-AAAAGGGACAGAAUGUGAGCAGUCACAAAUCUGCUGGCUUUUGGGCAUUCGGUUUUUUUUUUUUUUUUGUGUUCUUGCUGCUACAAUUUAUUUGUCUU --..........((.((((((-(((((((((.((((((.(((((........)))))..(....).)))))).)))))))))..............))))))))((((.....))))... ( -22.83) >DroYak_CAF1 61101 106 - 1 --CCUUUUUCUCGGUCGG-AA-AAAGGGGACAGAAUGUGAGCAGUCACAAAUCUGCUGGCUUUUGGGCAUUCGGUUU----------UUUGUGUUCUUGCUGCUACAAUUUAUUUGUCUU --(((((((((.....))-))-)))))(((((((...((((((((((((((.(((...(((....)))...)))...----------)))))).....)))))).)).....))))))). ( -34.10) >consensus __CCUUUUUCUCGGUCGGAAA_AAAAGGGACAGAAUGUGAGCAGUCACAAAUCUGCUGGCUUUUGGGCAUUCGGUUU__________UUUGUGCUCUGGCUGCUACAAUUUAUUUGUCUU ..((((((..((....))....))))))((((((.((((.(((((((((((.(((...(((....)))...))).............)))))).....))))))))).....)))))).. (-21.92 = -21.66 + -0.25)

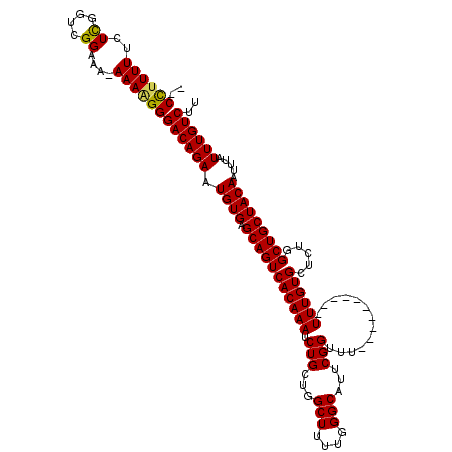
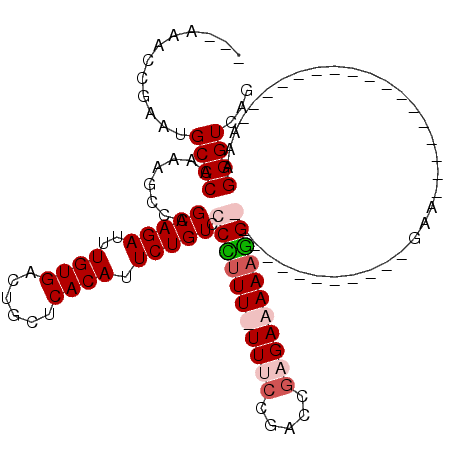
| Location | 2,943,795 – 2,943,893 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -19.31 |
| Energy contribution | -20.88 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2943795 98 + 22407834 ---AAACCGAAUGCCCAAAAGCCAGCAGAUUUGUGACUGCUCACAUUCUGUCCCUUUUUUUUCCGACCAAGAAAAAGGG--GGAAAUGGGGAA-----------------AAAGGGUCGG ---...((((...(((.....((.(((((..(((((....))))).)))))(((((((((((......)))))))))))--)).....)))..-----------------......)))) ( -32.60) >DroSim_CAF1 60666 117 + 1 ---AAACCGAAUGCCCAAAAGCCAGCAGAUUUGUGACUGCACACAUUCUGUCCCCUUUUUUUCCGACCGAGAAAAAGGGGGGGAAAUGGGGAAAAUGGGGGAAAGGGGUAAAAGGGUCGG ---...((((.(((((.....((.((((........))))...((((...(((((...((((((..((........))..)))))).))))).))))..))....)))))......)))) ( -40.30) >DroEre_CAF1 61059 90 + 1 AAAAAACCGAAUGCCCAAAAGCCAGCAGAUUUGUGACUGCUCACAUUCUGUCCCUUUU-UUGCCGACCGAGAAAAUAG------------GAA-----------------AAAGGGUCAG ............((((.....((.(((((..(((((....))))).)))))...((((-(((.....)))))))...)------------)..-----------------...))))... ( -19.70) >DroYak_CAF1 61134 86 + 1 ---AAACCGAAUGCCCAAAAGCCAGCAGAUUUGUGACUGCUCACAUUCUGUCCCCUUU-UU-CCGACCGAGAAAAAGG------------GAA-----------------AAAGGGUCAG ---.........((((........(((((..(((((....))))).))))).((((((-((-((....).))))))))------------)..-----------------...))))... ( -27.90) >consensus ___AAACCGAAUGCCCAAAAGCCAGCAGAUUUGUGACUGCUCACAUUCUGUCCCCUUU_UUUCCGACCGAGAAAAAGG____________GAA_________________AAAGGGUCAG ............((((........(((((..((((......)))).))))).(((((((((((.....)))))))))))..................................))))... (-19.31 = -20.88 + 1.56)

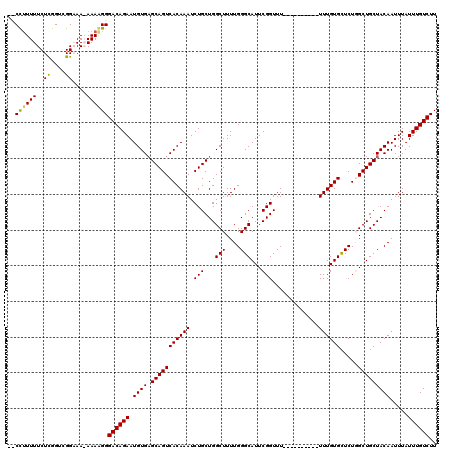
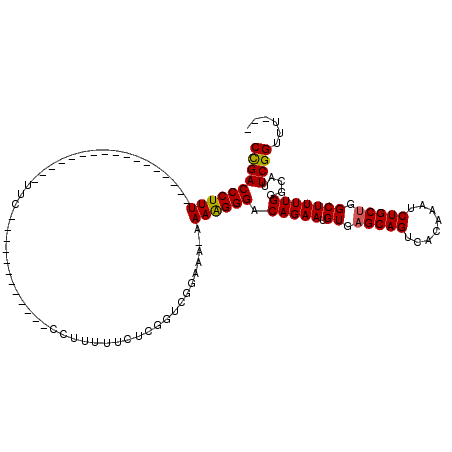
| Location | 2,943,795 – 2,943,893 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -19.62 |
| Energy contribution | -19.38 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2943795 98 - 22407834 CCGACCCUUU-----------------UUCCCCAUUUCC--CCCUUUUUCUUGGUCGGAAAAAAAAGGGACAGAAUGUGAGCAGUCACAAAUCUGCUGGCUUUUGGGCAUUCGGUUU--- ((((((((((-----------------((.....(((((--.((........))..))))))))))))).(((((.((.(((((........))))).))))))).....))))...--- ( -32.00) >DroSim_CAF1 60666 117 - 1 CCGACCCUUUUACCCCUUUCCCCCAUUUUCCCCAUUUCCCCCCCUUUUUCUCGGUCGGAAAAAAAGGGGACAGAAUGUGUGCAGUCACAAAUCUGCUGGCUUUUGGGCAUUCGGUUU--- (((((((.....((..................(((((...(((((((((.((.....)).)))))))))...)))))...((((........)))).)).....)))...))))...--- ( -29.90) >DroEre_CAF1 61059 90 - 1 CUGACCCUUU-----------------UUC------------CUAUUUUCUCGGUCGGCAA-AAAAGGGACAGAAUGUGAGCAGUCACAAAUCUGCUGGCUUUUGGGCAUUCGGUUUUUU ((((((((((-----------------(((------------(.(((.....))).))..)-))))))).(((((.((.(((((........))))).))))))).....))))...... ( -24.80) >DroYak_CAF1 61134 86 - 1 CUGACCCUUU-----------------UUC------------CCUUUUUCUCGGUCGG-AA-AAAGGGGACAGAAUGUGAGCAGUCACAAAUCUGCUGGCUUUUGGGCAUUCGGUUU--- ..((((....-----------------(((------------(((((((((.....))-))-))))))))(((((.((.(((((........))))).))))))).......)))).--- ( -30.90) >consensus CCGACCCUUU_________________UUC____________CCUUUUUCUCGGUCGGAAA_AAAAGGGACAGAAUGUGAGCAGUCACAAAUCUGCUGGCUUUUGGGCAUUCGGUUU___ ((((((((((.....................................................)))))).(((((.((.(((((........))))).))))))).....))))...... (-19.62 = -19.38 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:31 2006