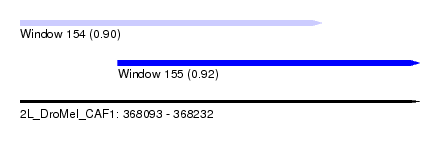
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 368,093 – 368,232 |
| Length | 139 |
| Max. P | 0.918006 |

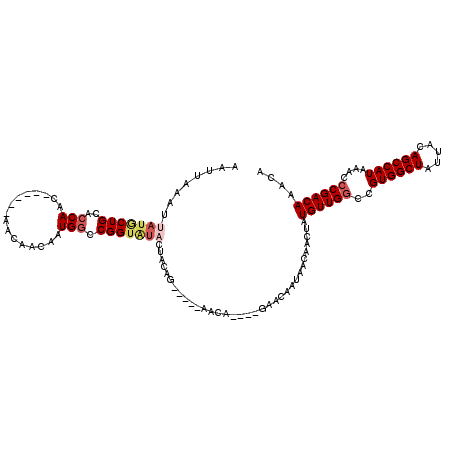
| Location | 368,093 – 368,198 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.40 |
| Mean single sequence MFE | -21.64 |
| Consensus MFE | -15.05 |
| Energy contribution | -16.21 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 368093 105 + 22407834 AAUUAAAUUAUGCUGCACCAAC------AACAACAAUGGCCGGUAUACUACGG-----AACA----GAACAAUAACAACUAUGUUGGCCGUGGCUAUUACAGCCAUAAACCCGACAAACA ........(((((((..(((..------........))).)))))))(....)-----....----...............((((((..((((((.....))))))....)))))).... ( -21.10) >DroSec_CAF1 35657 105 + 1 AAUUAAAUUAUACUGCACCAAC------AACAACAAUGGCCGGUAUACUACAG-----AACA----GAGCAAUAACAACUAUGUUGGCCGUGGCUAUUACAGCCAUAAACCCGACAAACA ........(((((((..(((..------........))).)))))))......-----....----...............((((((..((((((.....))))))....)))))).... ( -20.80) >DroSim_CAF1 36267 105 + 1 AAUUAAAUUAUGCUGCACCAAC------AACAACAAUGGCCGGUAUACUACAG-----AACA----GAGCAAUAACAACUAUGUUGGCCGUGGCUAUUACAGCCAUAAACCCGACAAACA ........(((((((..(((..------........))).)))))))......-----....----...............((((((..((((((.....))))))....)))))).... ( -20.80) >DroEre_CAF1 42110 102 + 1 AAUUAAAUUAUGCUGCACCAAC------ACCAACAAUGGCCGGG--------GCACAGAGCA----GAACAAUAACAACAAUGUUGUCCGUGGCUAUUACAGCCAUAAACCCGACAAACA ......................------.(((....))).((((--------((.....)).----(.((((((.......)))))).)((((((.....))))))...))))....... ( -19.50) >DroYak_CAF1 36683 120 + 1 AAUUAAAUUAUGCUGCACCAACACCAACACCAACAAUGCCCGGUGUUCUACAGCACAGAACAAUAACAACAAUAACAACAAUGUUGGCCGUGGCUAUUACAGCCAUAAACCCGACAAACA ..........(((((....((((((................))))))...)))))..........................((((((..((((((.....))))))....)))))).... ( -25.99) >consensus AAUUAAAUUAUGCUGCACCAAC______AACAACAAUGGCCGGUAUACUACAG_____AACA____GAACAAUAACAACUAUGUUGGCCGUGGCUAUUACAGCCAUAAACCCGACAAACA ........(((((((..(((................))).)))))))..................................((((((..((((((.....))))))....)))))).... (-15.05 = -16.21 + 1.16)

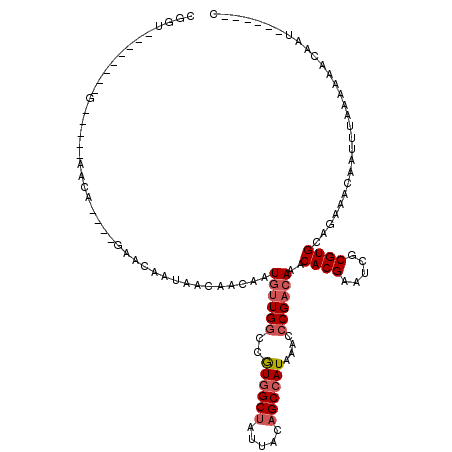
| Location | 368,127 – 368,232 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.36 |
| Mean single sequence MFE | -18.53 |
| Consensus MFE | -13.85 |
| Energy contribution | -14.47 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 368127 105 + 22407834 CGGUAUACUACGG-----AACA----GAACAAUAACAACUAUGUUGGCCGUGGCUAUUACAGCCAUAAACCCGACAAACACGAAUCGCGUGCAGAAACAAUUUAAAAAACAAU------C ((........)).-----....----...............((((((..((((((.....))))))....))))))..((((.....))))......................------. ( -18.00) >DroSim_CAF1 36301 105 + 1 CGGUAUACUACAG-----AACA----GAGCAAUAACAACUAUGUUGGCCGUGGCUAUUACAGCCAUAAACCCGACAAACACGAAUCGCGUGCAGAAACAAUUUAAAAAACAAU------C .............-----....----...............((((((..((((((.....))))))....))))))..((((.....))))......................------. ( -17.50) >DroEre_CAF1 42144 102 + 1 CGGG--------GCACAGAGCA----GAACAAUAACAACAAUGUUGUCCGUGGCUAUUACAGCCAUAAACCCGACAAACACGAAUCGCGUGCAGAAACAAUUUAAAAAACAAU------C ((((--------((.....)).----(.((((((.......)))))).)((((((.....))))))...)))).....((((.....))))......................------. ( -21.50) >DroWil_CAF1 75804 101 + 1 ------------------AACAGCAACAACAAUCAGAACAAU-UUGGUCAUGGCAAUUACAGCAAUAAAGCCGCCAAACACGAAUCGCGUGCAAAAACAAUUUAAAAAACAAUCAGAAUC ------------------.................((....(-(((((...(((...............)))))))))((((.....)))).....................))...... ( -11.16) >DroYak_CAF1 36723 114 + 1 CGGUGUUCUACAGCACAGAACAAUAACAACAAUAACAACAAUGUUGGCCGUGGCUAUUACAGCCAUAAACCCGACAAACACGAAUCGCGUGCAGAAACAAUUUAAAAAACAAU------C ...((((((.......))))))...................((((((..((((((.....))))))....))))))..((((.....))))......................------. ( -22.10) >DroAna_CAF1 47220 86 + 1 ----------------------------GCAAUAACAACAAUGUUGGCCAUGGCUAUUACAGCCAUAAAGCCGACAAACACGAAUCGCGUGCAGAAACAAUUUAAAAAACAAU------C ----------------------------.............(((((((.((((((.....))))))...)))))))..((((.....))))......................------. ( -20.90) >consensus CGGU________G_____AACA____GAACAAUAACAACAAUGUUGGCCGUGGCUAUUACAGCCAUAAACCCGACAAACACGAAUCGCGUGCAGAAACAAUUUAAAAAACAAU______C .........................................((((((..((((((.....))))))....))))))..((((.....))))............................. (-13.85 = -14.47 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:25:53 2006