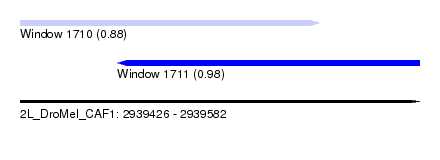
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,939,426 – 2,939,582 |
| Length | 156 |
| Max. P | 0.977321 |

| Location | 2,939,426 – 2,939,543 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.63 |
| Mean single sequence MFE | -30.51 |
| Consensus MFE | -25.91 |
| Energy contribution | -25.75 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2939426 117 + 22407834 CACAAAGGAUUUGG-GCAACA-GGAGGAAAUUGCAGCAAAUGUUAUGCCAUUUUAUUAAUGGAAUUUUUAUGUUGCACAUCCUUCUUGCU-GGAAUCUCAACGCUCUCACUUUAAGCGGC ......((((((.(-(((..(-((((((...(((((((.........(((((.....)))))........)))))))..)))))))))))-.))))))...((((.........)))).. ( -36.63) >DroSec_CAF1 54488 120 + 1 CACAAAGGAUUAGGAGGAACACGGAGGAAAUUGCAGCAAAUGUUAUGCCAUUUUAUUAAUGGAAUUUUUAUGUUGCACAUCCUUCUUGCUCGGAAUCUCAACGCUCUCGUUUUAAGCGCC ......(((((..(((.((...((((((...(((((((.........(((((.....)))))........)))))))..)))))))).)))..)))))...((((.........)))).. ( -30.13) >DroSim_CAF1 56532 120 + 1 CACAAAGGAUUAGGAGGAACACGGAGGAAAUUGCAGCAAAUGUUAUGCCAUUUUAUUAAUGGAAUUUUUAUGUUGCACAUCCUUCUUGCUCGGAAUCUCAACGCUCUCACUUUAAGCGCC ......(((((..(((.((...((((((...(((((((.........(((((.....)))))........)))))))..)))))))).)))..)))))...((((.........)))).. ( -30.13) >DroEre_CAF1 56779 117 + 1 CACAAAGGAUUAGU-GGAACA-GGAGGAAAUUGCAGCAAAUGUUAUGCCAUUUUAUUAAUGGAAUUUUUAUGUUGCACAUCCUUCUUGGU-GGAAUCUCAACGCUCUCACUUUAAGCGCC ......((...(((-(((.((-((((((...(((((((.........(((((.....)))))........)))))))..))).)))))((-(.........))))).)))).......)) ( -26.83) >DroYak_CAF1 56695 117 + 1 CACAAAGGAUUAGG-GGAACA-GAAGGAAAUUGCAGCAAAUGUUAUGCCAUUUUAUUAAUGGAAUUUUUAUGUUGCACAUCCUUCUUGGU-GGAAUCUCAACGCUCUCACUUUAAGCGCC ......((.(((((-(((..(-((((((...(((((((.........(((((.....)))))........)))))))..))))))).(((-(.........)))).)).))))))...)) ( -28.83) >consensus CACAAAGGAUUAGG_GGAACA_GGAGGAAAUUGCAGCAAAUGUUAUGCCAUUUUAUUAAUGGAAUUUUUAUGUUGCACAUCCUUCUUGCU_GGAAUCUCAACGCUCUCACUUUAAGCGCC ......(((((........((.((((((...(((((((.........(((((.....)))))........)))))))..)))))).)).....)))))...((((.........)))).. (-25.91 = -25.75 + -0.16)


| Location | 2,939,464 – 2,939,582 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.79 |
| Mean single sequence MFE | -35.14 |
| Consensus MFE | -26.84 |
| Energy contribution | -27.64 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2939464 118 - 22407834 GUGUCCGCUUAGCAUCCUUU-UUGGGGGCAAACAAAUUUCGCCGCUUAAAGUGAGAGCGUUGAGAUUCC-AGCAAGAAGGAUGUGCAACAUAAAAAUUCCAUUAAUAAAAUGGCAUAACA ((((..((...(((((((((-(((..((......(((((((.(((((.......))))).)))))))))-..)))))))))))))).)))).......(((((.....)))))....... ( -34.00) >DroSec_CAF1 54528 117 - 1 ---UCCGCAUAGCAUCCUUUUUUGGGGGCAAACAAAUUUCGGCGCUUAAAACGAGAGCGUUGAGAUUCCGAGCAAGAAGGAUGUGCAACAUAAAAAUUCCAUUAAUAAAAUGGCAUAACA ---...((...(((((((((((((((......).(((((((((((((.......)))))))))))))))))...))))))))))))............(((((.....)))))....... ( -32.50) >DroSim_CAF1 56572 119 - 1 GUGUCCGCAUAGCAUCCUUU-UUGGGGGCAAACAAAUUUCGGCGCUUAAAGUGAGAGCGUUGAGAUUCCGAGCAAGAAGGAUGUGCAACAUAAAAAUUCCAUUAAUAAAAUGGCAUAACA ((((..((...(((((((((-(((..((......(((((((((((((.......)))))))))))))))...)))))))))))))).)))).......(((((.....)))))....... ( -35.80) >DroEre_CAF1 56817 117 - 1 GUGUCUGCUGAGCAUCCUU--UUUGGGGCAAACAAAUUUGGGCGCUUAAAGUGAGAGCGUUGAGAUUCC-ACCAAGAAGGAUGUGCAACAUAAAAAUUCCAUUAAUAAAAUGGCAUAACA ((((.(((...((((((((--(((((((......(((((.(((((((.......))))))).)))))))-.)))))))))))))))))))).......(((((.....)))))....... ( -39.20) >DroYak_CAF1 56733 118 - 1 GUGUCUGCUUAGCAACCUUU-UUUGGGGCAAACAAAUUUCGGCGCUUAAAGUGAGAGCGUUGAGAUUCC-ACCAAGAAGGAUGUGCAACAUAAAAAUUCCAUUAAUAAAAUGGCAUAACA ((((.(((...(((.(((((-((.((((......(((((((((((((.......)))))))))))))))-.))))))))).)))))))))).......(((((.....)))))....... ( -34.20) >consensus GUGUCCGCUUAGCAUCCUUU_UUGGGGGCAAACAAAUUUCGGCGCUUAAAGUGAGAGCGUUGAGAUUCC_AGCAAGAAGGAUGUGCAACAUAAAAAUUCCAUUAAUAAAAUGGCAUAACA ......((...(((((((((......((......(((((((((((((.......)))))))))))))))......)))))))))))............(((((.....)))))....... (-26.84 = -27.64 + 0.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:24 2006