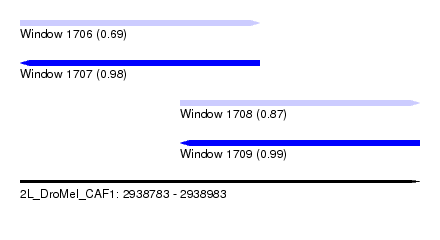
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,938,783 – 2,938,983 |
| Length | 200 |
| Max. P | 0.994799 |

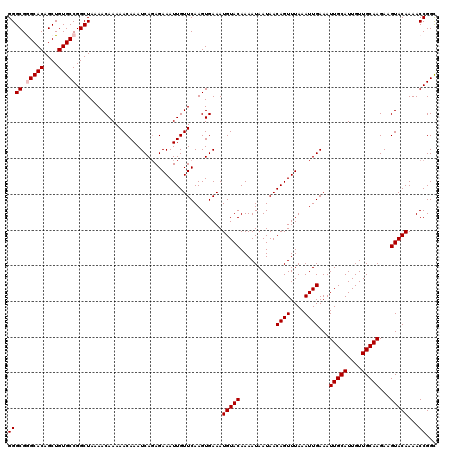
| Location | 2,938,783 – 2,938,903 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -21.81 |
| Energy contribution | -21.57 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2938783 120 + 22407834 GCCGGUUUUGUACUUCUUGCAACAAUGCAAUUUCAAUUUAAACUGUUAUUAUUUUGUACAUUUCACUUGAACAAUUUCUGUGAUUUGUUUUUGUUUUAGCCGGCACAGCUGUGCCCGCCC ..((((((........(((((....))))).........))))))............(((..((((..(((....))).))))..)))..........((.(((((....))))).)).. ( -25.43) >DroSec_CAF1 53824 120 + 1 GCCGGUUUUGUACUUCUUGCAACAAUGCAAUUUCAAUUUAAACUGUUAUUAUUUUGUACAUUUCACUUGAACAAUUUCUCUGAUUUGUUUUUGUUUUAGCCGGCACAGCUGUGCCCGCCC ..((((((........(((((....))))).........))))))...................((..((((((..........))))))..))....((.(((((....))))).)).. ( -22.33) >DroSim_CAF1 55862 120 + 1 GCCGGUUUUGUACUUCUUGCAACAAUGCAAUUUCAAUUUAAACUGUUAUUAUUUUGUACAUUUCACUUGAACAAUUUCUCUGAUUUGUUUUUGUUUUAGCCGGCACAGCUGUGCCCGCCC ..((((((........(((((....))))).........))))))...................((..((((((..........))))))..))....((.(((((....))))).)).. ( -22.33) >DroEre_CAF1 56142 120 + 1 ACCGGUUUUGUACUUCUUGCAACAAUGCAAUUUCAAUUUAAACUGUUAUUAUUUUGUACAUUUCACUUGAACAAUUUCUCUGAUUUGUUUUUGUUUUAGCCGGCACAGCUGUGCUCGCCC ..((((((........(((((....))))).........))))))...................((..((((((..........))))))..))....((.(((((....))))).)).. ( -20.43) >DroYak_CAF1 56041 120 + 1 GCCGGUUUUGUACUUCUUGCAACAAUGCAAUUUCAAUUUAAACUGUUAUUAUUUUGUACAUUUCACUUGAACAAUUUCUCUGAUUUGUUUUUGUUUUAGCCGGCACAGCUGUGCUCGCCC (((((((.(((((.....(((....)))(((..((........))..))).....)))))....((..((((((..........))))))..))...)))))))................ ( -22.00) >consensus GCCGGUUUUGUACUUCUUGCAACAAUGCAAUUUCAAUUUAAACUGUUAUUAUUUUGUACAUUUCACUUGAACAAUUUCUCUGAUUUGUUUUUGUUUUAGCCGGCACAGCUGUGCCCGCCC ..((((((........(((((....))))).........))))))...................((..((((((..........))))))..))....((.(((((....))))).)).. (-21.81 = -21.57 + -0.24)

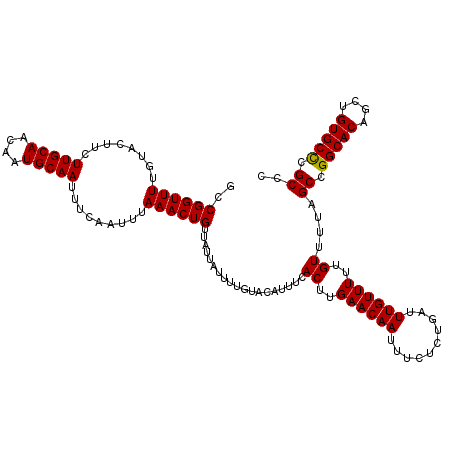
| Location | 2,938,783 – 2,938,903 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -23.38 |
| Energy contribution | -23.78 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2938783 120 - 22407834 GGGCGGGCACAGCUGUGCCGGCUAAAACAAAAACAAAUCACAGAAAUUGUUCAAGUGAAAUGUACAAAAUAAUAACAGUUUAAAUUGAAAUUGCAUUGUUGCAAGAAGUACAAAACCGGC ((((.(((((....))))).))...............((((.(((....)))..))))..(((((..........((((....))))...(((((....)))))...)))))...))... ( -28.60) >DroSec_CAF1 53824 120 - 1 GGGCGGGCACAGCUGUGCCGGCUAAAACAAAAACAAAUCAGAGAAAUUGUUCAAGUGAAAUGUACAAAAUAAUAACAGUUUAAAUUGAAAUUGCAUUGUUGCAAGAAGUACAAAACCGGC ((((.(((((....))))).))......................................(((((..........((((....))))...(((((....)))))...)))))...))... ( -26.10) >DroSim_CAF1 55862 120 - 1 GGGCGGGCACAGCUGUGCCGGCUAAAACAAAAACAAAUCAGAGAAAUUGUUCAAGUGAAAUGUACAAAAUAAUAACAGUUUAAAUUGAAAUUGCAUUGUUGCAAGAAGUACAAAACCGGC ((((.(((((....))))).))......................................(((((..........((((....))))...(((((....)))))...)))))...))... ( -26.10) >DroEre_CAF1 56142 120 - 1 GGGCGAGCACAGCUGUGCCGGCUAAAACAAAAACAAAUCAGAGAAAUUGUUCAAGUGAAAUGUACAAAAUAAUAACAGUUUAAAUUGAAAUUGCAUUGUUGCAAGAAGUACAAAACCGGU ((.((.((((....))))))(((..(((((................)))))..)))....(((((..........((((....))))...(((((....)))))...)))))...))... ( -22.09) >DroYak_CAF1 56041 120 - 1 GGGCGAGCACAGCUGUGCCGGCUAAAACAAAAACAAAUCAGAGAAAUUGUUCAAGUGAAAUGUACAAAAUAAUAACAGUUUAAAUUGAAAUUGCAUUGUUGCAAGAAGUACAAAACCGGC .(((.......)))..(((((......((..(((((..........)))))....))...(((((..........((((....))))...(((((....)))))...)))))...))))) ( -22.10) >consensus GGGCGGGCACAGCUGUGCCGGCUAAAACAAAAACAAAUCAGAGAAAUUGUUCAAGUGAAAUGUACAAAAUAAUAACAGUUUAAAUUGAAAUUGCAUUGUUGCAAGAAGUACAAAACCGGC ((((.(((((....))))).))......................................(((((..........((((....))))...(((((....)))))...)))))...))... (-23.38 = -23.78 + 0.40)

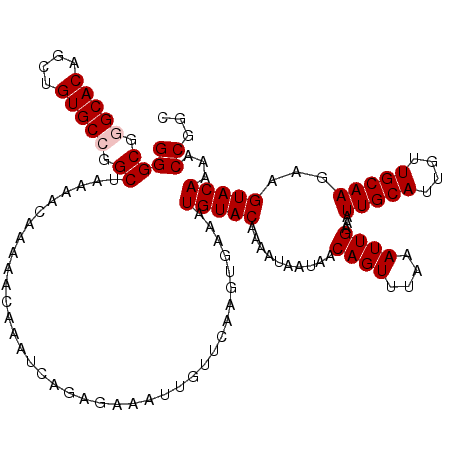
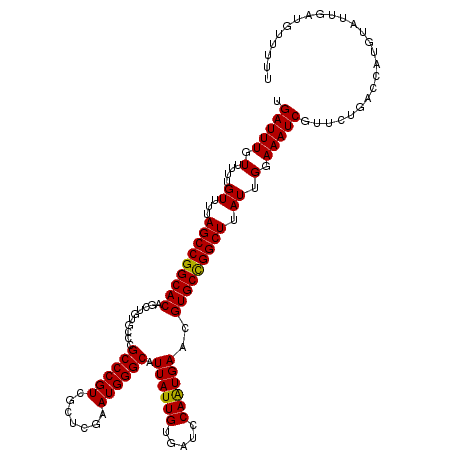
| Location | 2,938,863 – 2,938,983 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.66 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -33.42 |
| Energy contribution | -33.02 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.869842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2938863 120 + 22407834 UGAUUUGUUUUUGUUUUAGCCGGCACAGCUGUGCCCGCCCGUCGCUCGAAUGGGCAUUAUUGUGAUCCAGUGAACGUGCCGGCUUAUUGGAAAUCGUUCUGACCAUGUAUUGAUGUUUUU .((...((((..(....(((((((((.....((((((..((.....))..))))))((((((.....))))))..)))))))))...)..))))...))..................... ( -35.50) >DroSec_CAF1 53904 120 + 1 UGAUUUGUUUUUGUUUUAGCCGGCACAGCUGUGCCCGCCCGUCGCUCGAAUGGGCAUUAUUGUGAUCCAAUGAACGUGCCGGCUUAUUGGAAAUCGUUCUGACCAUGUAUUGAUGCUUUU .((...((((..(....(((((((((.....((((((..((.....))..))))))((((((.....))))))..)))))))))...)..))))...))..................... ( -35.50) >DroSim_CAF1 55942 120 + 1 UGAUUUGUUUUUGUUUUAGCCGGCACAGCUGUGCCCGCCCGUCGCUCGAAUGGGCAUUAUUGUGAUCCAAUGAACGUGCCGGCUUAUUGGAAAUCGUUCUGACCAUGUAUUGAUGUUUUU .((...((((..(....(((((((((.....((((((..((.....))..))))))((((((.....))))))..)))))))))...)..))))...))..................... ( -35.50) >DroEre_CAF1 56222 118 + 1 UGAUUUGUUUUUGUUUUAGCCGGCACAGCUGUGCUCGCCCGUCGCUUGAAUGGGCAUUAUUGUGAUCCAAUGAACGUGCUGGCUUAUUGGAAAUCGUUCUGACCAUGUAUUGAUAU--UU .((...((((..(....(((((((((..........((((((.......)))))).((((((.....))))))..)))))))))...)..))))...)).................--.. ( -31.90) >DroYak_CAF1 56121 118 + 1 UGAUUUGUUUUUGUUUUAGCCGGCACAGCUGUGCUCGCCCGUCGCUUCAAUGGGCAUUAUUGUGAUCCAAUGAACGUGCUGGCUUAUUGGAAAUCGUCCUGACCAUGUAUUGAUAU--UU .(((..((((..(....(((((((((..........((((((.......)))))).((((((.....))))))..)))))))))...)..)))).)))..................--.. ( -31.90) >consensus UGAUUUGUUUUUGUUUUAGCCGGCACAGCUGUGCCCGCCCGUCGCUCGAAUGGGCAUUAUUGUGAUCCAAUGAACGUGCCGGCUUAUUGGAAAUCGUUCUGACCAUGUAUUGAUGUUUUU .(((((.(....((...(((((((((..........((((((.......)))))).((((((.....))))))..))))))))).)).).)))))......................... (-33.42 = -33.02 + -0.40)

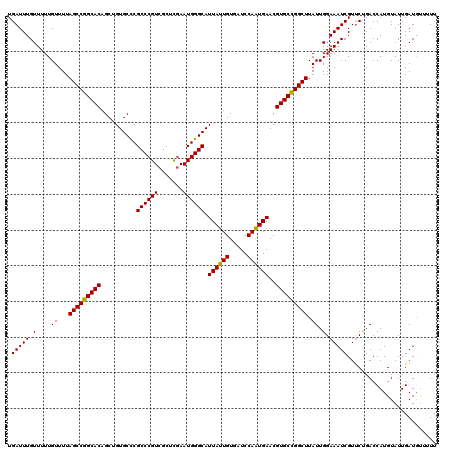
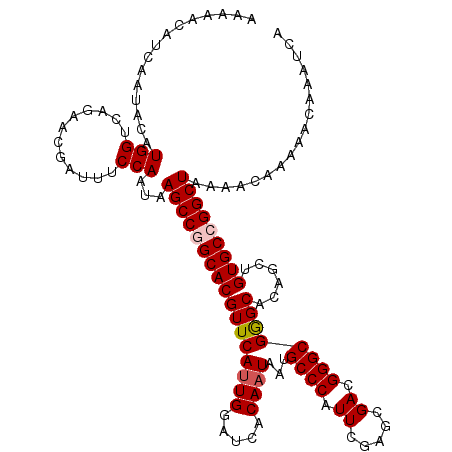
| Location | 2,938,863 – 2,938,983 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.66 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -26.38 |
| Energy contribution | -26.74 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2938863 120 - 22407834 AAAAACAUCAAUACAUGGUCAGAACGAUUUCCAAUAAGCCGGCACGUUCACUGGAUCACAAUAAUGCCCAUUCGAGCGACGGGCGGGCACAGCUGUGCCGGCUAAAACAAAAACAAAUCA ...............(((............)))...(((((((((......((.....))....(((((.((((.....)))).))))).....)))))))))................. ( -31.10) >DroSec_CAF1 53904 120 - 1 AAAAGCAUCAAUACAUGGUCAGAACGAUUUCCAAUAAGCCGGCACGUUCAUUGGAUCACAAUAAUGCCCAUUCGAGCGACGGGCGGGCACAGCUGUGCCGGCUAAAACAAAAACAAAUCA ...............(((............)))...(((((((((....((((.....))))..(((((.((((.....)))).))))).....)))))))))................. ( -33.20) >DroSim_CAF1 55942 120 - 1 AAAAACAUCAAUACAUGGUCAGAACGAUUUCCAAUAAGCCGGCACGUUCAUUGGAUCACAAUAAUGCCCAUUCGAGCGACGGGCGGGCACAGCUGUGCCGGCUAAAACAAAAACAAAUCA ...............(((............)))...(((((((((....((((.....))))..(((((.((((.....)))).))))).....)))))))))................. ( -33.20) >DroEre_CAF1 56222 118 - 1 AA--AUAUCAAUACAUGGUCAGAACGAUUUCCAAUAAGCCAGCACGUUCAUUGGAUCACAAUAAUGCCCAUUCAAGCGACGGGCGAGCACAGCUGUGCCGGCUAAAACAAAAACAAAUCA ..--...........(((((.(((....)))......((((((..((((((((.....))))...((((..((....)).))))))))...)))).)).)))))................ ( -24.50) >DroYak_CAF1 56121 118 - 1 AA--AUAUCAAUACAUGGUCAGGACGAUUUCCAAUAAGCCAGCACGUUCAUUGGAUCACAAUAAUGCCCAUUGAAGCGACGGGCGAGCACAGCUGUGCCGGCUAAAACAAAAACAAAUCA ..--...........(((((.(((.....))).....((((((..((((((((.....))))...((((.(((...))).))))))))...)))).)).)))))................ ( -28.40) >consensus AAAAACAUCAAUACAUGGUCAGAACGAUUUCCAAUAAGCCGGCACGUUCAUUGGAUCACAAUAAUGCCCAUUCGAGCGACGGGCGGGCACAGCUGUGCCGGCUAAAACAAAAACAAAUCA ...............(((............)))...(((((((((((((((((.....))))...((((.((.....)).))))))))......)))))))))................. (-26.38 = -26.74 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:22 2006