
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,937,848 – 2,937,951 |
| Length | 103 |
| Max. P | 0.753442 |

| Location | 2,937,848 – 2,937,951 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -33.72 |
| Consensus MFE | -25.16 |
| Energy contribution | -25.12 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2937848 103 + 22407834 -------------GGAUUCCGAUUCGGAUUCGCGCACAUGUGUUGCCAAACGCAGGGCUGCCAAGCAAAGCUAUCUGCUUGGGCAUAUACUAUAUAUG----UAUGUGUCGGAUACACAC -------------.((.((((...)))).))((((...((((((....))))))..)).))((((((........))))))(((((((((.......)----)))))))).......... ( -33.90) >DroSec_CAF1 52907 108 + 1 --------GAUUGGGAUUGGGAUUCGGAUUCGCGCACAUGUGUUGCCAAACGCAGGGCUGCCAAGCAAAGCUAUCUGCUUGGGCAUAUACUAUACAUA----UAUGUGGCGGAUACACAC --------.....((((....))))(.((((((.(((((((((.(((........)))(((((((((........))))).))))..........)))----)))))))))))).).... ( -32.60) >DroSim_CAF1 54909 107 + 1 -------------GGAUUGGGAUUCGGAUUCGCGCACAUGUGUUGCCAAACGCAGGACUGCCAAGCAAAGCUAUCUGCUUGGGCAUAUACUAUAUAUGUAUGUAUGUGGCGGAUACACAC -------------((((....))))(.((((((.(((((((.((((.....)))).))..(((((((........)))))))(((((((.......)))))))))))))))))).).... ( -33.20) >DroEre_CAF1 55195 104 + 1 AGGAUGAAGAUGAGGGUUCGGAUUCGGAUUCGCGCACAUGUGUUGCCAAAUGCAGCGCUGCCAAGCAAAGCUAUCUGCUUGGGCAUAU---------------AUGU-GCGGAUACACAC .............(((((((....))))))).(((((((((((.(((....(((....)))((((((........)))))))))))))---------------))))-)))......... ( -36.80) >DroYak_CAF1 55056 97 + 1 --------GAUGAGAAUUCGGAUUUGGAUUCGCGCACAUGUGUUGCCAAACGCAGAGCUGCCAAGCAAAGCUAUCUGCUUGGGCAUAU---------------AUGUAGCGGAUACACAC --------.....(((((((....))))))).(((((((((((.(((....(((....)))((((((........)))))))))))))---------------)))).)))......... ( -32.10) >consensus ________GAU__GGAUUCGGAUUCGGAUUCGCGCACAUGUGUUGCCAAACGCAGGGCUGCCAAGCAAAGCUAUCUGCUUGGGCAUAUACUAUA_AU_____UAUGUGGCGGAUACACAC .............((((....))))(.((((((.(((((((.((((.....)))).))(((((((((........))))).))))..................))))))))))).).... (-25.16 = -25.12 + -0.04)

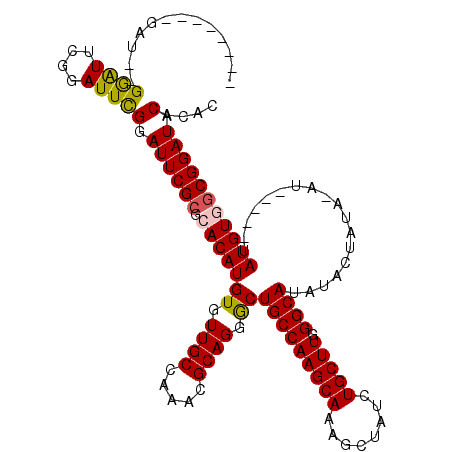
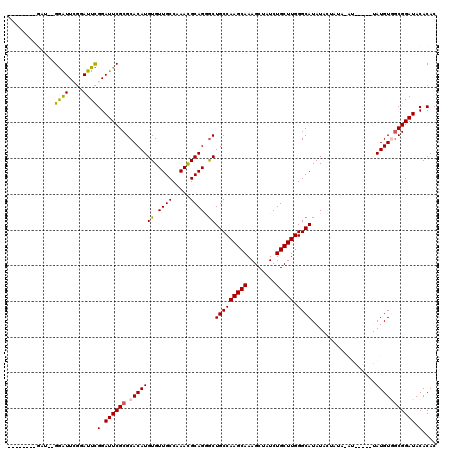
| Location | 2,937,848 – 2,937,951 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -28.94 |
| Consensus MFE | -22.54 |
| Energy contribution | -22.38 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2937848 103 - 22407834 GUGUGUAUCCGACACAUA----CAUAUAUAGUAUAUGCCCAAGCAGAUAGCUUUGCUUGGCAGCCCUGCGUUUGGCAACACAUGUGCGCGAAUCCGAAUCGGAAUCC------------- ((((((.....)))))).----........(((((((.(((((((((....)))))))))..(((........)))....)))))))..((.((((...)))).)).------------- ( -31.70) >DroSec_CAF1 52907 108 - 1 GUGUGUAUCCGCCACAUA----UAUGUAUAGUAUAUGCCCAAGCAGAUAGCUUUGCUUGGCAGCCCUGCGUUUGGCAACACAUGUGCGCGAAUCCGAAUCCCAAUCCCAAUC-------- .........((((((((.----..(((........((((.(((((((....)))))))))))(((........))).))).))))).)))......................-------- ( -26.90) >DroSim_CAF1 54909 107 - 1 GUGUGUAUCCGCCACAUACAUACAUAUAUAGUAUAUGCCCAAGCAGAUAGCUUUGCUUGGCAGUCCUGCGUUUGGCAACACAUGUGCGCGAAUCCGAAUCCCAAUCC------------- .........((((((((..((((.......)))).((((((((((((....))))))))(((....)))....))))....))))).))).................------------- ( -28.00) >DroEre_CAF1 55195 104 - 1 GUGUGUAUCCGC-ACAU---------------AUAUGCCCAAGCAGAUAGCUUUGCUUGGCAGCGCUGCAUUUGGCAACACAUGUGCGCGAAUCCGAAUCCGAACCCUCAUCUUCAUCCU (((((....)))-))..---------------...((((.(((((((....)))))))))))((((.((((.((....)).))))))))............................... ( -32.20) >DroYak_CAF1 55056 97 - 1 GUGUGUAUCCGCUACAU---------------AUAUGCCCAAGCAGAUAGCUUUGCUUGGCAGCUCUGCGUUUGGCAACACAUGUGCGCGAAUCCAAAUCCGAAUUCUCAUC-------- (((((((.....)))))---------------)).((((.(((((((....)))))))))))((.(.((((.((....)).))))).))((((.(......).)))).....-------- ( -25.90) >consensus GUGUGUAUCCGCCACAUA_____AU_UAUAGUAUAUGCCCAAGCAGAUAGCUUUGCUUGGCAGCCCUGCGUUUGGCAACACAUGUGCGCGAAUCCGAAUCCGAAUCC__AUC________ .(((((.............................((((.(((((((....))))))))))).....((((.((....)).))))))))).............................. (-22.54 = -22.38 + -0.16)

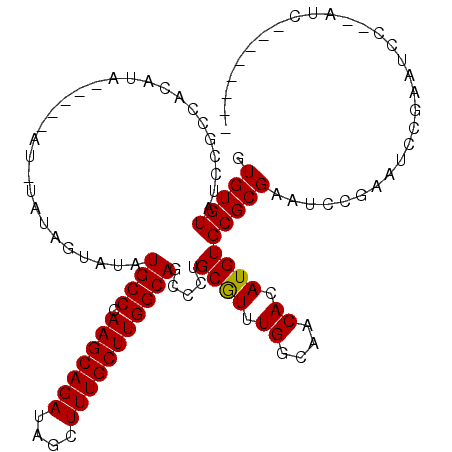
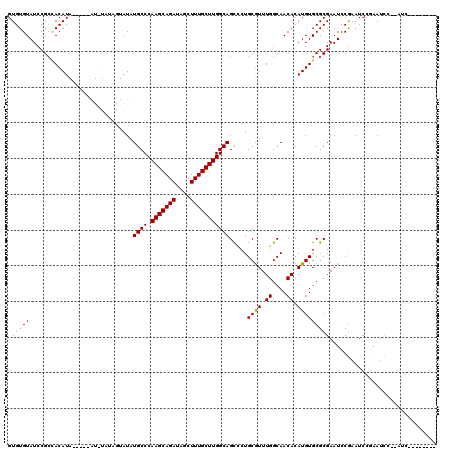
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:17 2006