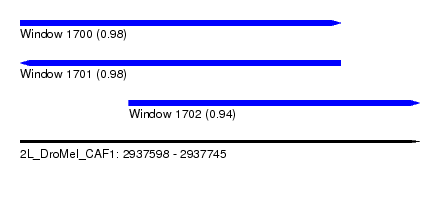
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,937,598 – 2,937,745 |
| Length | 147 |
| Max. P | 0.982534 |

| Location | 2,937,598 – 2,937,716 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.48 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -22.04 |
| Energy contribution | -22.44 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.977037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2937598 118 + 22407834 GCACGAAUAUGGAGCCAUAAAAGAGCAUCUACCGCAAAUCGCUUAUAUCACUCAGCAGCGCCUAAAAGCGGGCAACGAA-AAACAAUUUAAAGUUACAUCGACAACUUUAAUGGCCCAA- .........(((.(((((.((((........((((....((((.............)))).......))))....(((.-.(((........)))...)))....)))).)))))))).- ( -24.32) >DroSec_CAF1 52671 118 + 1 ACACGAAUAUGGAGCCAUAAAAGAGCAUGUACCGCAAAUCGCUUAUAUCACUCAGCAGCGCCUAAAAGCGGGCAACGAA-AAACAAUUUAAAGUUACAUCGACAACUUUAAUGGCCCAA- .........(((.(((((.((((....(((.((((....((((.............)))).......))))))).(((.-.(((........)))...)))....)))).)))))))).- ( -25.42) >DroSim_CAF1 54674 118 + 1 ACACGAAUAUGGAGCCAUAAAAGAGCAUGUACCGCAAAUCGCUUAUAUCACUCAGCAGCGCCUAAAAGCGGGCAACGAA-AAACAAUUUAAAGUUACAUCGACAACUUUAAUGGCCCAA- .........(((.(((((.((((....(((.((((....((((.............)))).......))))))).(((.-.(((........)))...)))....)))).)))))))).- ( -25.42) >DroEre_CAF1 54954 119 + 1 ACACGAAUAUGGAGCCAUAAAAAAGCAUGUACCGCAAAUCGCUUAUAUCACUCAGCAGCGCCUAAAAGCGGGCAACGAAAAAACAAUUUGAAGUUACAUCGACAACUUUAAUGGCCCAA- .........(((.(((((.........(((.((((....((((.............)))).......)))))))..............(((((((........))))))))))))))).- ( -24.02) >DroYak_CAF1 54827 120 + 1 ACACGAAUAUGGACCCAUAAAAGAGCAUGUACCGCAAAUCGCUUAUAUCACUCAGCAGCGCCUAAAAGCGGGCAACGAAAAAACAAUUUAAAGUUACAUCGACAACUUUAAUGGCCCAAA .........(((..((((.((((....(((.((((....((((.............)))).......))))))).(((...(((........)))...)))....)))).)))).))).. ( -20.62) >consensus ACACGAAUAUGGAGCCAUAAAAGAGCAUGUACCGCAAAUCGCUUAUAUCACUCAGCAGCGCCUAAAAGCGGGCAACGAA_AAACAAUUUAAAGUUACAUCGACAACUUUAAUGGCCCAA_ .........(((.(((((.((((........((((....((((.............)))).......))))....(((...(((........)))...)))....)))).)))))))).. (-22.04 = -22.44 + 0.40)


| Location | 2,937,598 – 2,937,716 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.48 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -31.00 |
| Energy contribution | -31.08 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2937598 118 - 22407834 -UUGGGCCAUUAAAGUUGUCGAUGUAACUUUAAAUUGUUU-UUCGUUGCCCGCUUUUAGGCGCUGCUGAGUGAUAUAAGCGAUUUGCGGUAGAUGCUCUUUUAUGGCUCCAUAUUCGUGC -.((((((((((((((((......)))))))))...((((-.((((....(((((.((..((((....)))).)).)))))....)))).)))).........)))).)))......... ( -30.20) >DroSec_CAF1 52671 118 - 1 -UUGGGCCAUUAAAGUUGUCGAUGUAACUUUAAAUUGUUU-UUCGUUGCCCGCUUUUAGGCGCUGCUGAGUGAUAUAAGCGAUUUGCGGUACAUGCUCUUUUAUGGCUCCAUAUUCGUGU -.((((((((.((((..((..(((((.((.((((((((((-.(((..((.((((....))))..))....)))...)))))))))).))))))))).)))).))))).)))......... ( -33.00) >DroSim_CAF1 54674 118 - 1 -UUGGGCCAUUAAAGUUGUCGAUGUAACUUUAAAUUGUUU-UUCGUUGCCCGCUUUUAGGCGCUGCUGAGUGAUAUAAGCGAUUUGCGGUACAUGCUCUUUUAUGGCUCCAUAUUCGUGU -.((((((((.((((..((..(((((.((.((((((((((-.(((..((.((((....))))..))....)))...)))))))))).))))))))).)))).))))).)))......... ( -33.00) >DroEre_CAF1 54954 119 - 1 -UUGGGCCAUUAAAGUUGUCGAUGUAACUUCAAAUUGUUUUUUCGUUGCCCGCUUUUAGGCGCUGCUGAGUGAUAUAAGCGAUUUGCGGUACAUGCUUUUUUAUGGCUCCAUAUUCGUGU -.((((((((.(((((.....(((((.((.((((((((((..(((..((.((((....))))..))....)))...)))))))))).))))))))))))...))))).)))......... ( -34.20) >DroYak_CAF1 54827 120 - 1 UUUGGGCCAUUAAAGUUGUCGAUGUAACUUUAAAUUGUUUUUUCGUUGCCCGCUUUUAGGCGCUGCUGAGUGAUAUAAGCGAUUUGCGGUACAUGCUCUUUUAUGGGUCCAUAUUCGUGU ..((((((...((((..((..(((((.((.((((((((((..(((..((.((((....))))..))....)))...)))))))))).))))))))).))))....))))))......... ( -31.50) >consensus _UUGGGCCAUUAAAGUUGUCGAUGUAACUUUAAAUUGUUU_UUCGUUGCCCGCUUUUAGGCGCUGCUGAGUGAUAUAAGCGAUUUGCGGUACAUGCUCUUUUAUGGCUCCAUAUUCGUGU ..((((((((.((((..((..(((((.((.((((((((((..(((..((.((((....))))..))....)))...)))))))))).))))))))).)))).))))).)))......... (-31.00 = -31.08 + 0.08)


| Location | 2,937,638 – 2,937,745 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -23.22 |
| Consensus MFE | -22.68 |
| Energy contribution | -23.08 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2937638 107 + 22407834 GCUUAUAUCACUCAGCAGCGCCUAAAAGCGGGCAACGAA-AAACAAUUUAAAGUUACAUCGACAACUUUAAUGGCCCAA-AGGGCAA-AAAGAUGGCCAUUACGAUGAGG ..........((((.(..(((......)))(((......-.......((((((((........))))))))..((((..-.))))..-.......))).....).)))). ( -23.30) >DroSec_CAF1 52711 107 + 1 GCUUAUAUCACUCAGCAGCGCCUAAAAGCGGGCAACGAA-AAACAAUUUAAAGUUACAUCGACAACUUUAAUGGCCCAA-AGGGCAA-AAAGAUGGCCAUUACGAGGAGC ((((...((.........(((......)))(((......-.......((((((((........))))))))..((((..-.))))..-.......))).....)).)))) ( -23.30) >DroSim_CAF1 54714 105 + 1 GCUUAUAUCACUCAGCAGCGCCUAAAAGCGGGCAACGAA-AAACAAUUUAAAGUUACAUCGACAACUUUAAUGGCCCAA-AGGGCAA-AAAGAUGGCCAUUACGAGGA-- ..........(((.....(((......)))(((......-.......((((((((........))))))))..((((..-.))))..-.......))).....)))..-- ( -22.30) >DroEre_CAF1 54994 106 + 1 GCUUAUAUCACUCAGCAGCGCCUAAAAGCGGGCAACGAAAAAACAAUUUGAAGUUACAUCGACAACUUUAAUGGCCCAA-AGGGCAA-AAAGAUGGCCAUUAGGACGA-- (((..........)))..(((((((....((((...(......)...((((((((........))))))))..))))..-.((.((.-.....)).)).))))).)).-- ( -23.10) >DroYak_CAF1 54867 108 + 1 GCUUAUAUCACUCAGCAGCGCCUAAAAGCGGGCAACGAAAAAACAAUUUAAAGUUACAUCGACAACUUUAAUGGCCCAAAAGGGCAAAAAAGAUGGCCAUUAGGAGGA-- ..........(((.....(((......)))(((..(((...(((........)))...))).((.((((....((((....))))...)))).))))).....)))..-- ( -24.10) >consensus GCUUAUAUCACUCAGCAGCGCCUAAAAGCGGGCAACGAA_AAACAAUUUAAAGUUACAUCGACAACUUUAAUGGCCCAA_AGGGCAA_AAAGAUGGCCAUUACGAGGA__ ..........(((.....(((......)))(((..(((...(((........)))...))).((.((((....((((....))))...)))).))))).....))).... (-22.68 = -23.08 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:15 2006