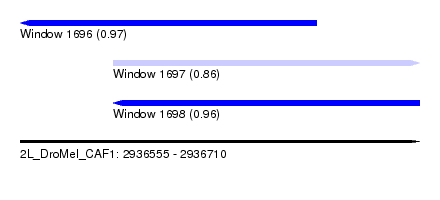
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,936,555 – 2,936,710 |
| Length | 155 |
| Max. P | 0.968439 |

| Location | 2,936,555 – 2,936,670 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -20.41 |
| Consensus MFE | -17.90 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2936555 115 - 22407834 UUAAUCAAUUAAUUUCAAUUAAAUUGUGAAAAAUCACAACAACGUGAGG-AAAAACAAGCGCAACAAACAUGUUUAUGGUUUCCGUUGUGCACAGAAAACUGCAAA----UUAAGUAUAU ........(((((((((......((((......((((......))))..-....))))(((((((((((.........))))..))))))).........)).)))----))))...... ( -19.90) >DroSec_CAF1 51964 119 - 1 UUAAUCAAUUAAUUUCAAUUAAAUUGUGAAAAAUCACAACAACGUGAGG-AAAAACAAGCGCAACAAACAUGUUUAUGGUUUCCGUUGUGCACAGAAAACUCCAAAGGAAAUAAAUAUAU .......(((.(((((.......((((......((((......))))..-....))))(((((((((((.........))))..)))))))................))))).))).... ( -20.00) >DroSim_CAF1 53643 119 - 1 UUAAUCAAUUAAUUUCAAUUAAAUUGUGAAAAAUCACAACAACGUGAGG-AAAAACAAGCGCAACAAACAUGUUUAUGGUUUCCGUUGUGCGCAGAAAACUCCAAAGGAAAUAAAUAUAU .......(((.(((((.......((((((....))))))....(.(((.-........(((((((((((.........))))..)))))))........))))....))))).))).... ( -20.73) >DroEre_CAF1 53942 114 - 1 UUAAUCAAUUAAUUUCAAUUAAAUUGUGAAAAAUCACAACAACGUGAGGUGAAAACCAGCGCAACAAACAUGUCUAUGGUUUCCGUUGUGCACAGAAAACUACAAAGGUAAUA------U ............((((.......((((((....))))))....((..(((....))).(((((((((((.........))))..))))))))).))))...............------. ( -21.70) >DroYak_CAF1 53848 113 - 1 UUAAUCAAUUAAUUUCAAUUAAAUUGUGAAAAAUCACAACAACGUGAGG-AAAAACAAGCGCAACAAACAUGUUUAUGGUUUCCGUUGUGCAAAGAAAACUCCAAGGGAAAUA------U ...........(((((.......((((......((((......))))..-....))))(((((((((((.........))))..)))))))................))))).------. ( -19.70) >consensus UUAAUCAAUUAAUUUCAAUUAAAUUGUGAAAAAUCACAACAACGUGAGG_AAAAACAAGCGCAACAAACAUGUUUAUGGUUUCCGUUGUGCACAGAAAACUCCAAAGGAAAUAA_UAUAU ............((((.......((((((....))))))...(....)..........(((((((((((.........))))..)))))))...))))...................... (-17.90 = -17.90 + 0.00)

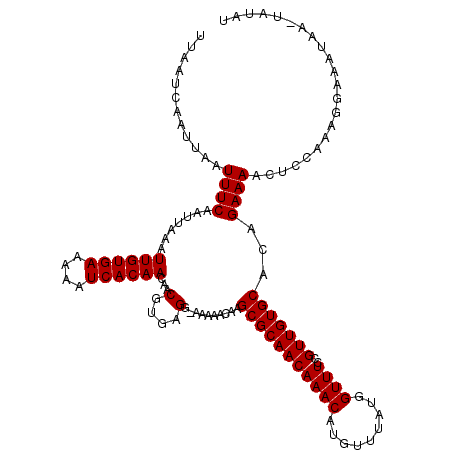
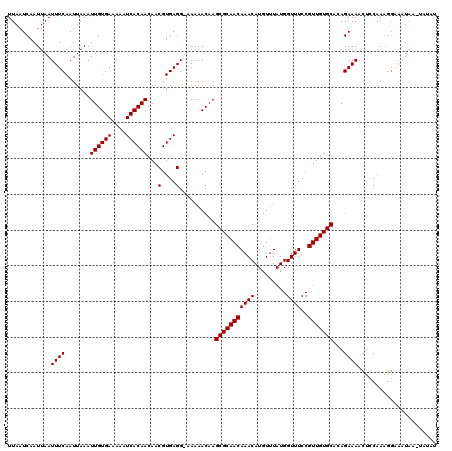
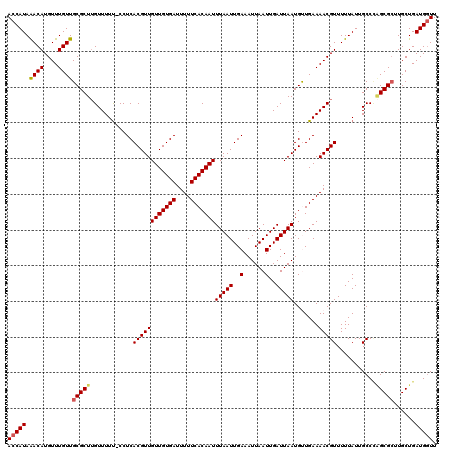
| Location | 2,936,591 – 2,936,710 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -25.41 |
| Consensus MFE | -22.54 |
| Energy contribution | -22.86 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2936591 119 + 22407834 ACCAUAAACAUGUUUGUUGCGCUUGUUUUU-CCUCACGUUGUUGUGAUUUUUCACAAUUUAAUUGAAAUUAAUUGAUUAAUGUUGAAAACGUUUUUAUUGCCCAGCGCUUGCUGAUGAUU ..(((.((((....))))(((((.((....-....((((((((((((....)))))))(((((..(......)..))))).......))))).......))..)))))......)))... ( -21.96) >DroSec_CAF1 52004 119 + 1 ACCAUAAACAUGUUUGUUGCGCUUGUUUUU-CCUCACGUUGUUGUGAUUUUUCACAAUUUAAUUGAAAUUAAUUGAUUAAUGUUGAAAACGUUUUUAUUGCCCAGCGCUUGCUGAUGGUU (((((.((((....))))(((((.((....-....((((((((((((....)))))))(((((..(......)..))))).......))))).......))..)))))......))))). ( -25.66) >DroSim_CAF1 53683 119 + 1 ACCAUAAACAUGUUUGUUGCGCUUGUUUUU-CCUCACGUUGUUGUGAUUUUUCACAAUUUAAUUGAAAUUAAUUGAUUAAUGUUGAAAACGUUUUUAUUGCCCAGCGAUUGCUGAUGGUU (((((...((...((((((.((.(((((((-....((((((((((((....)))))).(((((((....))))))).)))))).)))))))........)).)))))).))...))))). ( -24.40) >DroEre_CAF1 53976 120 + 1 ACCAUAGACAUGUUUGUUGCGCUGGUUUUCACCUCACGUUGUUGUGAUUUUUCACAAUUUAAUUGAAAUUAAUUGAUUAAUGCUGAAAACGUUUUUAUUGCCAGGCGCUUGCUGAUGGUU (((((.((((....))))((((((((.........((((((((((((....)))))))(((((..(......)..))))).......))))).......)))).))))......))))). ( -30.09) >DroYak_CAF1 53882 119 + 1 ACCAUAAACAUGUUUGUUGCGCUUGUUUUU-CCUCACGUUGUUGUGAUUUUUCACAAUUUAAUUGAAAUUAAUUGAUUAAUGCUGAAAACGUUUUUAUUGCCUUGCGCUUGCUGAUGGUU (((((.((((....))))((((..((....-....((((((((((((....)))))))(((((..(......)..))))).......))))).......))...))))......))))). ( -24.96) >consensus ACCAUAAACAUGUUUGUUGCGCUUGUUUUU_CCUCACGUUGUUGUGAUUUUUCACAAUUUAAUUGAAAUUAAUUGAUUAAUGUUGAAAACGUUUUUAUUGCCCAGCGCUUGCUGAUGGUU (((((.((((....))))(((((............((((((((((((....)))))))(((((..(......)..))))).......)))))...........)))))......))))). (-22.54 = -22.86 + 0.32)

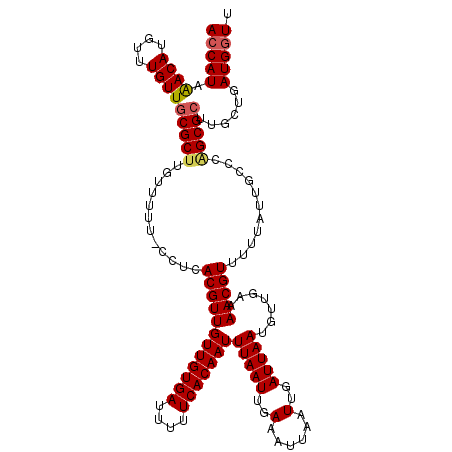
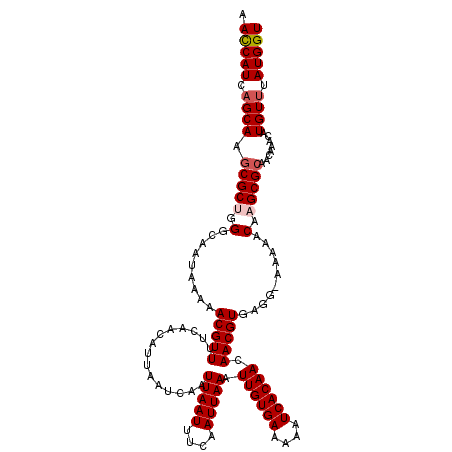
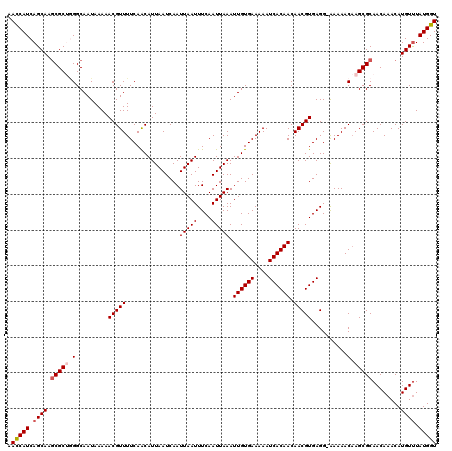
| Location | 2,936,591 – 2,936,710 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -20.52 |
| Consensus MFE | -18.13 |
| Energy contribution | -18.77 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2936591 119 - 22407834 AAUCAUCAGCAAGCGCUGGGCAAUAAAAACGUUUUCAACAUUAAUCAAUUAAUUUCAAUUAAAUUGUGAAAAAUCACAACAACGUGAGG-AAAAACAAGCGCAACAAACAUGUUUAUGGU .(((((.((((.(((((...........(((((...............(((((....))))).((((((....)))))).)))))....-.......)))))........)))).))))) ( -19.25) >DroSec_CAF1 52004 119 - 1 AACCAUCAGCAAGCGCUGGGCAAUAAAAACGUUUUCAACAUUAAUCAAUUAAUUUCAAUUAAAUUGUGAAAAAUCACAACAACGUGAGG-AAAAACAAGCGCAACAAACAUGUUUAUGGU .(((((.((((.(((((...........(((((...............(((((....))))).((((((....)))))).)))))....-.......)))))........)))).))))) ( -21.85) >DroSim_CAF1 53683 119 - 1 AACCAUCAGCAAUCGCUGGGCAAUAAAAACGUUUUCAACAUUAAUCAAUUAAUUUCAAUUAAAUUGUGAAAAAUCACAACAACGUGAGG-AAAAACAAGCGCAACAAACAUGUUUAUGGU .(((((.((((...(.((.((.........((.....)).............((((.......((((((....))))))...(....))-))).....)).)).).....)))).))))) ( -18.30) >DroEre_CAF1 53976 120 - 1 AACCAUCAGCAAGCGCCUGGCAAUAAAAACGUUUUCAGCAUUAAUCAAUUAAUUUCAAUUAAAUUGUGAAAAAUCACAACAACGUGAGGUGAAAACCAGCGCAACAAACAUGUCUAUGGU .(((((..(((.((.....)).........((((...((...............(((......((((((....)))))).....)))(((....)))...))...)))).)))..))))) ( -23.20) >DroYak_CAF1 53882 119 - 1 AACCAUCAGCAAGCGCAAGGCAAUAAAAACGUUUUCAGCAUUAAUCAAUUAAUUUCAAUUAAAUUGUGAAAAAUCACAACAACGUGAGG-AAAAACAAGCGCAACAAACAUGUUUAUGGU .(((((.((((.((((...((...(((....)))...)).............((((.......((((((....))))))...(....))-))).....))))........)))).))))) ( -20.00) >consensus AACCAUCAGCAAGCGCUGGGCAAUAAAAACGUUUUCAACAUUAAUCAAUUAAUUUCAAUUAAAUUGUGAAAAAUCACAACAACGUGAGG_AAAAACAAGCGCAACAAACAUGUUUAUGGU .(((((.((((.(((((.(.........(((((...............(((((....))))).((((((....)))))).)))))..........).)))))........)))).))))) (-18.13 = -18.77 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:12 2006