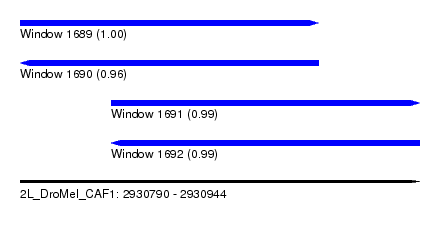
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,930,790 – 2,930,944 |
| Length | 154 |
| Max. P | 0.996797 |

| Location | 2,930,790 – 2,930,905 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.21 |
| Mean single sequence MFE | -43.22 |
| Consensus MFE | -31.76 |
| Energy contribution | -32.12 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2930790 115 + 22407834 CAACACACCCACCCCUCCUGCAGUGAUCCC-CU---UUCAACCGACUUGACUGGAUGUUUUGCUCAAUAUGCCAGUCAGUCACUUGGCCGGGAAAACAUGGCCAAAAGCGGUGGGGCGG ........((.((((.((.((.........-..---.......((((.(((((((((((......))))).))))))))))..(((((((........)))))))..)))).)))).)) ( -43.70) >DroSec_CAF1 48521 109 + 1 --------CCACCCCUCCU-CAGUGAUCCC-CCCUGCUCAACCGACUUGACUGGAUGUUUGGCUCAAUAUGCCAGUCAGUCACUUGGCCGGGAAAACAUGGCCAAAAGCGGUGGGGUGG --------(((((((.((.-.((.(.....-.)))(((.....((((.(((((((((((......))))).))))))))))..(((((((........))))))).))))).))))))) ( -45.60) >DroSim_CAF1 50104 110 + 1 --------CCACCCCUCCU-CAGUGAUCCCACCCUGCUCAACCGACUUGACUGGAUGUUUUGCUCAAUAUGCCAGUCAGUCACUUGGCCGGGAAAACAUGGCCAAAAGCGGUGGGGUGG --------(((((((.((.-..(((....)))...(((.....((((.(((((((((((......))))).))))))))))..(((((((........))))))).))))).))))))) ( -48.60) >DroEre_CAF1 50190 105 + 1 CACC--AUCCACCCGACUG-CCGUGAUCCC-CU---UUCAACCGACUUGACUGGAUGUUUUGCUCAAUAUGCCAGUCAGUCACUUGGCCGAGAAAACAUGGCCAGAAA--GGGG----- ....--...(((.......-..)))..(((-((---(((....((((.(((((((((((......))))).))))))))))...((((((.(....).))))))))))--))))----- ( -39.00) >DroYak_CAF1 50120 107 + 1 CACC--ACCCACCCCACCA-CUGUGAUCCC-CU---UUCAACCGACUUGACUGGAUGUUUUGCUCAAUAUGCCAGUCAGUCACUUGGCCGGGAAAACAUGGCCAGAAGUGGGGG----- ..((--.(((((....(((-.(((..((((-..---.((((..((((.(((((((((((......))))).))))))))))..))))..))))..))))))......)))))))----- ( -39.20) >consensus CA_C__A_CCACCCCUCCU_CAGUGAUCCC_CU___UUCAACCGACUUGACUGGAUGUUUUGCUCAAUAUGCCAGUCAGUCACUUGGCCGGGAAAACAUGGCCAAAAGCGGUGGGG_GG ........(((((..............................((((.(((((((((((......))))).))))))))))..(((((((........)))))))....)))))..... (-31.76 = -32.12 + 0.36)

| Location | 2,930,790 – 2,930,905 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.21 |
| Mean single sequence MFE | -46.66 |
| Consensus MFE | -29.58 |
| Energy contribution | -31.90 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2930790 115 - 22407834 CCGCCCCACCGCUUUUGGCCAUGUUUUCCCGGCCAAGUGACUGACUGGCAUAUUGAGCAAAACAUCCAGUCAAGUCGGUUGAA---AG-GGGAUCACUGCAGGAGGGGUGGGUGUGUUG .(((((((((.((((((.(..((..(((((...(((.(((((((((((.((.((......)).)))))))).))))).)))..---.)-)))).))..))))))).)))))).)))... ( -50.00) >DroSec_CAF1 48521 109 - 1 CCACCCCACCGCUUUUGGCCAUGUUUUCCCGGCCAAGUGACUGACUGGCAUAUUGAGCCAAACAUCCAGUCAAGUCGGUUGAGCAGGG-GGGAUCACUG-AGGAGGGGUGG-------- (((((((.((((.....))((((((..(((.(((((.(((((((((((.((.((......)).)))))))).))))).))).)).)))-..)).)).))-.)).)))))))-------- ( -50.40) >DroSim_CAF1 50104 110 - 1 CCACCCCACCGCUUUUGGCCAUGUUUUCCCGGCCAAGUGACUGACUGGCAUAUUGAGCAAAACAUCCAGUCAAGUCGGUUGAGCAGGGUGGGAUCACUG-AGGAGGGGUGG-------- (((((((.((((.....))((.((..((((((((((.(((((((((((.((.((......)).)))))))).))))).))).))....)))))..))))-.)).)))))))-------- ( -46.70) >DroEre_CAF1 50190 105 - 1 -----CCCC--UUUCUGGCCAUGUUUUCUCGGCCAAGUGACUGACUGGCAUAUUGAGCAAAACAUCCAGUCAAGUCGGUUGAA---AG-GGGAUCACGG-CAGUCGGGUGGAU--GGUG -----((((--((((.((((..(....)..))))((.(((((((((((.((.((......)).)))))))).))))).)))))---))-)))(((((.(-(......)).).)--))). ( -39.40) >DroYak_CAF1 50120 107 - 1 -----CCCCCACUUCUGGCCAUGUUUUCCCGGCCAAGUGACUGACUGGCAUAUUGAGCAAAACAUCCAGUCAAGUCGGUUGAA---AG-GGGAUCACAG-UGGUGGGGUGGGU--GGUG -----((((((((((..(((((...(((((...(((.(((((((((((.((.((......)).)))))))).))))).)))..---.)-)))).....)-)))))))))))).--)).. ( -46.80) >consensus CC_CCCCACCGCUUUUGGCCAUGUUUUCCCGGCCAAGUGACUGACUGGCAUAUUGAGCAAAACAUCCAGUCAAGUCGGUUGAA___AG_GGGAUCACUG_AGGAGGGGUGG_U__G_UG (((((((.((......((((..........))))((.(((((((((((.((.((......)).)))))))).))))).)).....................)).)))))))........ (-29.58 = -31.90 + 2.32)


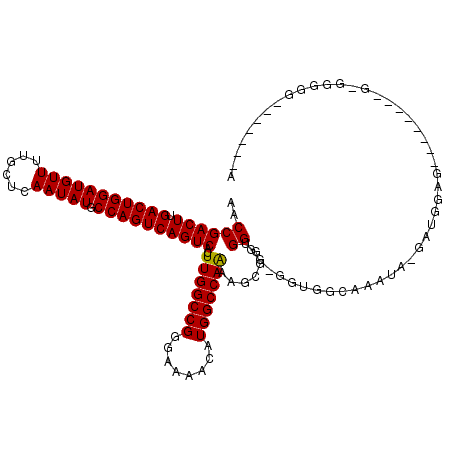
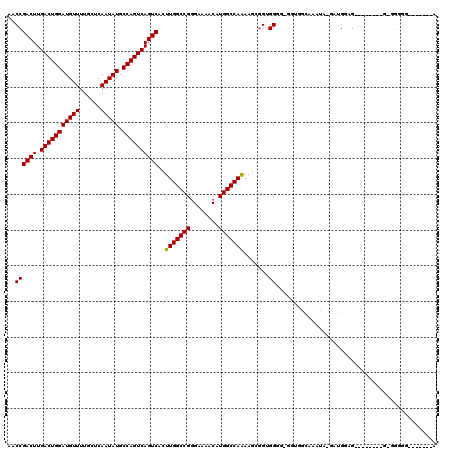
| Location | 2,930,825 – 2,930,944 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.74 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -28.48 |
| Energy contribution | -28.24 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2930825 119 + 22407834 AACCGACUUGACUGGAUGUUUUGCUCAAUAUGCCAGUCAGUCACUUGGCCGGGAAAACAUGGCCAAAAGCGGUGGGGCGGUGGCAAACA-GAAGGAGAGUGGAGGGGGGGGGGGUUGGUA .((((((((..((...(.(((..(((....((((((((...((((((((((........)))))).....)))).)))..)))))..(.-...)..)))..))).).))..)))))))). ( -39.60) >DroSec_CAF1 48550 96 + 1 AACCGACUUGACUGGAUGUUUGGCUCAAUAUGCCAGUCAGUCACUUGGCCGGGAAAACAUGGCCAAAAGCGGUGGGGUGGUGGCAAGUG-CUAGUAG----------------------- .(((((((.(((((((((((......))))).))))))))))..(((((((........)))))))....)))....((.((((....)-))).)).----------------------- ( -34.90) >DroSim_CAF1 50134 96 + 1 AACCGACUUGACUGGAUGUUUUGCUCAAUAUGCCAGUCAGUCACUUGGCCGGGAAAACAUGGCCAAAAGCGGUGGGGUGGUGGCAAGUG-GUUGUAG----------------------- ((((((((.(((((((((((......))))).))))))))))(((((.(((..(...(((.((.....)).)))...)..)))))))))-)))....----------------------- ( -34.00) >DroEre_CAF1 50222 99 + 1 AACCGACUUGACUGGAUGUUUUGCUCAAUAUGCCAGUCAGUCACUUGGCCGAGAAAACAUGGCCAGAAA--GGGG-------ACAAAUA-GAUGGAG-------CGAGGGGG----GCAA ..((((((.(((((((((((......))))).))))))))))..(((((((.(....).)))))))...--(...-------.).....-...)).(-------(.......----)).. ( -30.00) >DroYak_CAF1 50152 100 + 1 AACCGACUUGACUGGAUGUUUUGCUCAAUAUGCCAGUCAGUCACUUGGCCGGGAAAACAUGGCCAGAAGUGGGGG-------AGAAAUAAAAUGGAG-------CGAGGGGG------UA .(((..((((((((((((((......))))).))))))..(((((((((((........))))))..)))))...-------...............-------.)))..))------). ( -31.10) >consensus AACCGACUUGACUGGAUGUUUUGCUCAAUAUGCCAGUCAGUCACUUGGCCGGGAAAACAUGGCCAAAAGCGGUGGGG_GGUGGCAAAUA_GAUGGAG________G_GGGGG_______A ..((((((.(((((((((((......))))).))))))))))..(((((((........))))))).......))............................................. (-28.48 = -28.24 + -0.24)

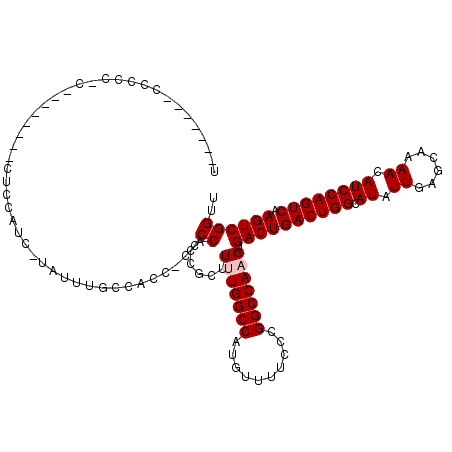
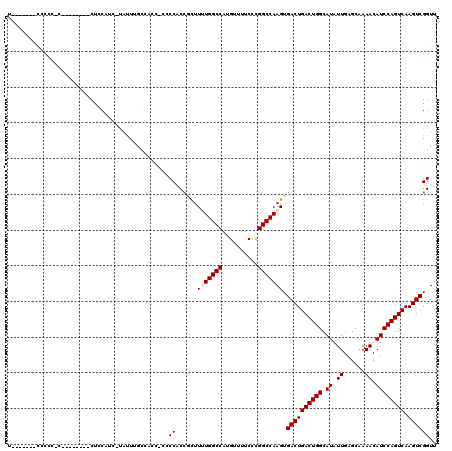
| Location | 2,930,825 – 2,930,944 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.74 |
| Mean single sequence MFE | -24.96 |
| Consensus MFE | -22.72 |
| Energy contribution | -23.12 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2930825 119 - 22407834 UACCAACCCCCCCCCCCUCCACUCUCCUUC-UGUUUGCCACCGCCCCACCGCUUUUGGCCAUGUUUUCCCGGCCAAGUGACUGACUGGCAUAUUGAGCAAAACAUCCAGUCAAGUCGGUU ..............................-.....(((...((......)).(((((((..........))))))).((((((((((.((.((......)).)))))))).))))))). ( -25.60) >DroSec_CAF1 48550 96 - 1 -----------------------CUACUAG-CACUUGCCACCACCCCACCGCUUUUGGCCAUGUUUUCCCGGCCAAGUGACUGACUGGCAUAUUGAGCCAAACAUCCAGUCAAGUCGGUU -----------------------......(-(....)).........(((...(((((((..........))))))).((((((((((.((.((......)).)))))))).))))))). ( -26.00) >DroSim_CAF1 50134 96 - 1 -----------------------CUACAAC-CACUUGCCACCACCCCACCGCUUUUGGCCAUGUUUUCCCGGCCAAGUGACUGACUGGCAUAUUGAGCAAAACAUCCAGUCAAGUCGGUU -----------------------....(((-(.....................(((((((..........))))))).((((((((((.((.((......)).)))))))).)))))))) ( -26.20) >DroEre_CAF1 50222 99 - 1 UUGC----CCCCCUCG-------CUCCAUC-UAUUUGU-------CCCC--UUUCUGGCCAUGUUUUCUCGGCCAAGUGACUGACUGGCAUAUUGAGCAAAACAUCCAGUCAAGUCGGUU ..((----(.......-------.......-.......-------....--....(((((..(....)..)))))...((((((((((.((.((......)).)))))))).))))))). ( -23.40) >DroYak_CAF1 50152 100 - 1 UA------CCCCCUCG-------CUCCAUUUUAUUUCU-------CCCCCACUUCUGGCCAUGUUUUCCCGGCCAAGUGACUGACUGGCAUAUUGAGCAAAACAUCCAGUCAAGUCGGUU .(------((......-------...............-------.....(((..(((((..........))))))))((((((((((.((.((......)).)))))))).))))))). ( -23.60) >consensus U_______CCCCC_C________CUCCAUC_UAUUUGCCACC_CCCCACCGCUUUUGGCCAUGUUUUCCCGGCCAAGUGACUGACUGGCAUAUUGAGCAAAACAUCCAGUCAAGUCGGUU .............................................((......(((((((..........))))))).((((((((((.((.((......)).)))))))).)))))).. (-22.72 = -23.12 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:06 2006