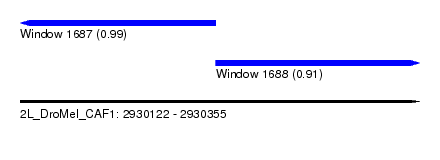
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,930,122 – 2,930,355 |
| Length | 233 |
| Max. P | 0.986365 |

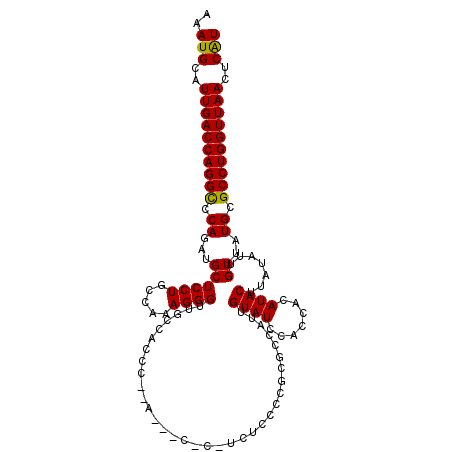
| Location | 2,930,122 – 2,930,236 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 84.82 |
| Mean single sequence MFE | -29.89 |
| Consensus MFE | -22.54 |
| Energy contribution | -22.22 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2930122 114 - 22407834 AAAUGCAUUGACCAGGCCCAGAUGCUCCUGCCAAAGGGUUCGCACCACCA--CCACCACUCCCCGCGCCAUUGUAUCCACCACAUACUUAUAUUGUUAUGCGCCUGGUUAACUCAU ..(((..((((((((((.(((((((....((....(((............--.........)))..))....)))))....(((.........)))..)).))))))))))..))) ( -25.60) >DroSec_CAF1 47895 109 - 1 AAAUGCAUUGACCAGGCCCAGAUGCUCCUGCCAAAGGGUUGCUACCACCA-------GCUCCCCUCGCCAUUGUAUCCACCACAUACUUAUAUUGUUAUGCGCCUGGUUAACUCAU ..(((..((((((((((.(((((((....((...((((..(((......)-------))..)))).))....)))))....(((.........)))..)).))))))))))..))) ( -32.80) >DroSim_CAF1 49494 103 - 1 AAAUGCAUUGACCAGGCCCAGAUGCUCCUGCCAAAGGGUUGCUACUC-------------CUUCUCGCCAUUGUAUCCACCACAUACUUAUAUUGUUAUGCGCCUGGUUAACUCAU ..(((..((((((((((.(((((((....((..((((((....)).)-------------)))...))....)))))....(((.........)))..)).))))))))))..))) ( -27.40) >DroEre_CAF1 49516 116 - 1 AAAUGCAUUGACCAGGCCCAGAUGCUCCUGCCAAAGGGUUCCCACCCAAACGUCCCUUGUUGCUGAAGCAUUGUAUCCACCACAUACUUAUAUUGUUAUGCGCCUGGUUAACUCGU ..(((..((((((((((.((((((((.(.((..(((((...............)))))...)).).))))))((((.......))))...........)).))))))))))..))) ( -32.76) >DroYak_CAF1 49447 111 - 1 AAAUGCAUUGACCAGGUCCAGAUGCUCCUGCCAAAGGGUUGCCCCUC-----CCCCCUGUUGCUGCAGCAUUGUAUCCACCACAUACUUAUAUUGUUAUGCGCCUGGUUAACUCAU ..(((..((((((((((.((((((((.(.((...((((.........-----..))))...)).).))))))((((.......))))...........)).))))))))))..))) ( -30.90) >consensus AAAUGCAUUGACCAGGCCCAGAUGCUCCUGCCAAAGGGUUGCCACCC__A___C_C_UCUCCCCGCGCCAUUGUAUCCACCACAUACUUAUAUUGUUAUGCGCCUGGUUAACUCAU ..(((..((((((((((.((...((((((.....))))..................................((((.......)))).......))..)).))))))))))..))) (-22.54 = -22.22 + -0.32)

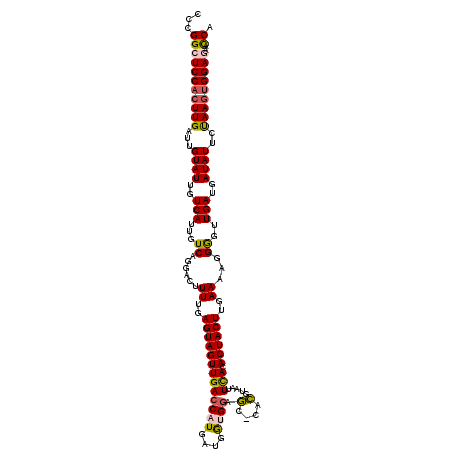
| Location | 2,930,236 – 2,930,355 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.55 |
| Mean single sequence MFE | -36.56 |
| Consensus MFE | -27.78 |
| Energy contribution | -27.66 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2930236 119 + 22407834 CCCGGCUCCACUUGAUUGUAUUGUCAUUGUCAGGACUUUUGAGUAGUUGACCAUGAUGGUGGAGC-CACGUAAUUCACACUACUUGAAAAGGGGUUGAUGAUAUUCUAAGUGGAGAACCA ...(((((((((((...((((..(((...((....(((((((((((((((..(((.(((.....)-)))))...))).))))))).))))).)).)))..))))..)))))))))..)). ( -37.70) >DroSec_CAF1 48004 119 + 1 CCCGGCUCCACUUGAUUGUAUUGUCAUUGUCAGGACUUUUGAGUAGUUGACCAUGAUGGUGGAGC-CACGUAAUUCACACUACUUGAAAAGGAGUUGAUGAUAUUCUAAGUGGACAACCA ...((.((((((((...((((..(((...((.....(((..(((((((((..(((.(((.....)-)))))...))).))))))..)))..))..)))..))))..))))))))...)). ( -35.50) >DroSim_CAF1 49597 119 + 1 CCCGGCUCCACUUGAUUGUAUUGUCAUUGUCAGGACUUUUGAGUAGUUGACCAUGAUGGUGGAGC-CACGUAAUUCACACUACUUGAAAAGGGGUUGAUGAUAUUCUAAGUGGAGAACCA ...(((((((((((...((((..(((...((....(((((((((((((((..(((.(((.....)-)))))...))).))))))).))))).)).)))..))))..)))))))))..)). ( -37.70) >DroEre_CAF1 49632 119 + 1 CCCGGCUCCACUUGAUUGUAUUGUCAUUGUCAGGACUUUUGAGUAGUUGACCAUGCUCAAGGACUGUGGGUAAUUUACACUACUUGAA-AGGGCUUGAUGAUAUUCCAAGUGGAGAAUCA .....(((((((((...((((..(((..(((....((((..((((((......((((((.......))))))......))))))..))-))))).)))..))))..)))))))))..... ( -41.60) >DroYak_CAF1 49558 119 + 1 CCCGGCUCCACUUGAUUGUAUUGUCAUUGUCAGGACUUUUGAGUAGUUGACCAUGCUCAAGCACUGUGGGUAAUUUACACUACUUCAA-AGGGGUUGAUGAUAUUCCAAAAGGAAAACCA ..(((((((.((((((.((......)).))))))...((((((((((......((((((.......))))))......))))).))))-))))))))......((((....))))..... ( -30.30) >consensus CCCGGCUCCACUUGAUUGUAUUGUCAUUGUCAGGACUUUUGAGUAGUUGACCAUGAUGGUGGAGC_CACGUAAUUCACACUACUUGAAAAGGGGUUGAUGAUAUUCUAAGUGGAGAACCA ...(((((((((((...((((..(((...((......((..(((((((((((((....)))).(....).....))).))))))..))...))..)))..))))..)))))))))..)). (-27.78 = -27.66 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:02 2006