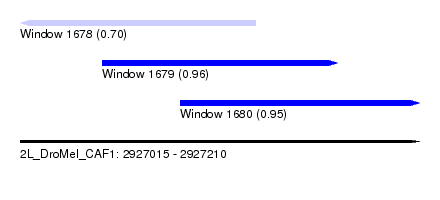
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,927,015 – 2,927,210 |
| Length | 195 |
| Max. P | 0.959617 |

| Location | 2,927,015 – 2,927,130 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.21 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -17.32 |
| Energy contribution | -17.56 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2927015 115 - 22407834 ---ACGCCCAUUCAGGGCCAUCCCAUUAGCAAAUACAACAGCGGCUAUAAUUAUAAGCCACUCCCUCCGCUCACCC--UCUCCCUUCUUCUCCACAGAAUAAAUGGCUAACUUUGUUGAU ---..((((.....))))......(((((((((......(((((((.........)))).................--.........((((....))))......)))...))))))))) ( -19.50) >DroSec_CAF1 45269 112 - 1 ---ACGCCCAUUCAGGGCCAUCCCAUUAGCAAAUACAACAGCGGCUAUAAUUAUAAGCCAUCCCCUUCGCUCACCCCCUCUCCC---GACGCCACAGAAUAAAUGGCUAACUUUGUUG-- ---..((((.....)))).................((((((.((((.........)))).........................---...((((.........)))).....))))))-- ( -19.90) >DroSim_CAF1 46391 99 - 1 ---ACGCCCAUUCAGGGCCAUCCCAUUAGCAAAUACAACAGCGGCUAUAAUUAUAAGCC-------------ACCCCCUCUCCC---GUCGCCACAGAAUAAAUGGCUAACUUUGUUG-- ---..((((.....)))).................((((((.((((.........))))-------------............---...((((.........)))).....))))))-- ( -19.90) >DroEre_CAF1 46331 108 - 1 ---ACGCCCAUUCAGGGCCAUCCCAUUAGCAAAUACAACAGCGGCUAUAAUUAUAAGCCC-C---CCCGCUCGCCC--UCUCCCGG-CU--CCGCAGAAUAAAUGGCUAACUUUGUUGAU ---..((((.....))))......(((((((((......(((((((.........)))).-.---...((..(((.--......))-).--..))..........)))...))))))))) ( -23.30) >DroYak_CAF1 46258 114 - 1 ACGACGCCCAUUCAGGGCCAUCCCAUUAGCAAAUACAACAGCGGCUAUAAUUAUAAGCCCUC---CCCUCUCACCC--UCUCCCGU-AUCGCCACGGAAUAAAUGGCUAACUUUGUUGAU .((((((((.....)))).......(((((.........((.((((.........)))))).---...........--....((((-......))))........)))))....)))).. ( -24.30) >consensus ___ACGCCCAUUCAGGGCCAUCCCAUUAGCAAAUACAACAGCGGCUAUAAUUAUAAGCCA_C___CCCGCUCACCC__UCUCCC___GUCGCCACAGAAUAAAUGGCUAACUUUGUUGAU .....((((.....)))).................((((((.((((.........))))...............................((((.........)))).....)))))).. (-17.32 = -17.56 + 0.24)

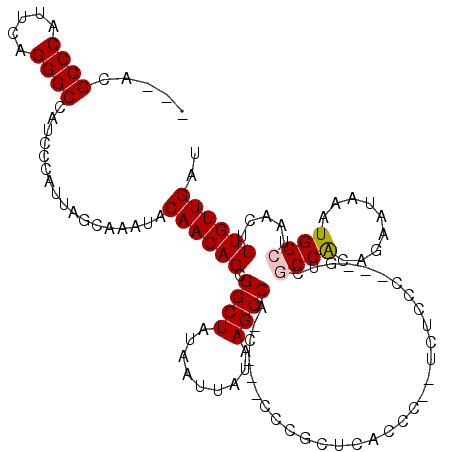
| Location | 2,927,055 – 2,927,170 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.82 |
| Mean single sequence MFE | -40.86 |
| Consensus MFE | -33.64 |
| Energy contribution | -34.08 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2927055 115 + 22407834 GA--GGGUGAGCGGAGGGAGUGGCUUAUAAUUAUAGCCGCUGUUGUAUUUGCUAAUGGGAUGGCCCUGAAUGGGCGU---CGUUCGGGGCGUGGCAAUUAAAGCACCUUGUCCAAGUGCU ..--(((..((.(.....(((((((.........)))))))(((.((.((((((........((((((((((.....---)))))))))).)))))).)).))).)))..)))....... ( -40.70) >DroSec_CAF1 45304 117 + 1 GAGGGGGUGAGCGAAGGGGAUGGCUUAUAAUUAUAGCCGCUGUUGUAUUUGCUAAUGGGAUGGCCCUGAAUGGGCGU---CGUUCGGGGCGUGGCAAUUAAAGCACCUUGUCCAAGUGCU ((.(((((((((.(......).)))..........(((((.(((.((((....)))).))).((((((((((.....---)))))))))))))))........)))))).))........ ( -39.90) >DroSim_CAF1 46426 104 + 1 GAGGGGGU-------------GGCUUAUAAUUAUAGCCGCUGUUGUAUUUGCUAAUGGGAUGGCCCUGAAUGGGCGU---CGUUCGGGGCGUGGCAAUUAAAGCACCUUGUCCAAGUGCU ((.(((((-------------(.(((.(((((...(((((.(((.((((....)))).))).((((((((((.....---))))))))))))))))))))))))))))).))........ ( -40.10) >DroEre_CAF1 46368 111 + 1 GA--GGGCGAGCGGG---G-GGGCUUAUAAUUAUAGCCGCUGUUGUAUUUGCUAAUGGGAUGGCCCUGAAUGGGCGU---CGUUCGGGGCGUGGCAAUUAAAGCACCUUGUCCAAGUGCU .(--(.((..(((((---(-..((((.(((((...(((((.(((.((((....)))).))).((((((((((.....---)))))))))))))))))))))))).))))))....)).)) ( -42.40) >DroYak_CAF1 46297 115 + 1 GA--GGGUGAGAGGG---GAGGGCUUAUAAUUAUAGCCGCUGUUGUAUUUGCUAAUGGGAUGGCCCUGAAUGGGCGUCGUCGUUCGGGGCGUGGCAAUUAAAGCACCUUGUCCAAGUGCU ..--.((((...(((---((((((((.(((((...(((((.(((.((((....)))).))).(((((((((((......))))))))))))))))))))))))).)))).)))...)))) ( -41.20) >consensus GA__GGGUGAGCGGA___G_GGGCUUAUAAUUAUAGCCGCUGUUGUAUUUGCUAAUGGGAUGGCCCUGAAUGGGCGU___CGUUCGGGGCGUGGCAAUUAAAGCACCUUGUCCAAGUGCU ....(((((((..........((((.........))))(((....((.((((((........((((((((((........)))))))))).)))))).)).)))..)))))))....... (-33.64 = -34.08 + 0.44)

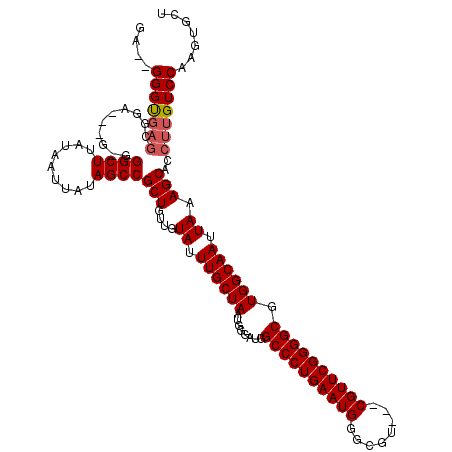
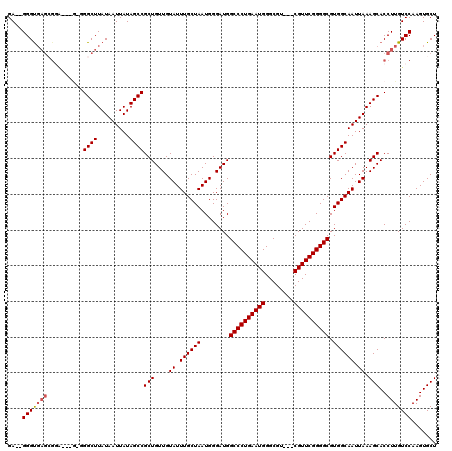
| Location | 2,927,093 – 2,927,210 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.80 |
| Mean single sequence MFE | -42.02 |
| Consensus MFE | -38.62 |
| Energy contribution | -38.66 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2927093 117 + 22407834 UGUUGUAUUUGCUAAUGGGAUGGCCCUGAAUGGGCGU---CGUUCGGGGCGUGGCAAUUAAAGCACCUUGUCCAAGUGCUGCGAUUCCGGGCGGCGUGCAAUCAGUUGAAUUGCAAUUGG .((((((.((((((......)))).((((.((.((((---(((((((((((..(((......((.....)).....)))..)).))))))))))))).)).))))...)).))))))... ( -44.30) >DroSec_CAF1 45344 117 + 1 UGUUGUAUUUGCUAAUGGGAUGGCCCUGAAUGGGCGU---CGUUCGGGGCGUGGCAAUUAAAGCACCUUGUCCAAGUGCUGCGAUUUCGGGCGGCGUGCAAUCAGUUGAAUUGCAAUUGG .((((((.((((((......)))).((((.((.((((---((((((..(((..(((......((.....)).....)))..)).)..)))))))))).)).))))...)).))))))... ( -42.50) >DroSim_CAF1 46453 117 + 1 UGUUGUAUUUGCUAAUGGGAUGGCCCUGAAUGGGCGU---CGUUCGGGGCGUGGCAAUUAAAGCACCUUGUCCAAGUGCUGCGAUUUCGGGCGGCGUGCAAUCAGUUGAAUUGCAAUUGG .((((((.((((((......)))).((((.((.((((---((((((..(((..(((......((.....)).....)))..)).)..)))))))))).)).))))...)).))))))... ( -42.50) >DroEre_CAF1 46402 117 + 1 UGUUGUAUUUGCUAAUGGGAUGGCCCUGAAUGGGCGU---CGUUCGGGGCGUGGCAAUUAAAGCACCUUGUCCAAGUGCUGCGAUUUCGGCCGGAGUGCAAUCAGUUGAAUUGCAAUUGG .(((((((((............((((((((((.....---)))))))))).((((......(((((.((....))))))).((....)))))))))))))))((((((.....)))))). ( -39.30) >DroYak_CAF1 46332 120 + 1 UGUUGUAUUUGCUAAUGGGAUGGCCCUGAAUGGGCGUCGUCGUUCGGGGCGUGGCAAUUAAAGCACCUUGUCCAAGUGCUGCGAUUUCGGGCGGAGUGCAAUCAGUUGAAUUGCAAUUGG .((((((((((((...(..((.(((((((((((......)))))))))))(..(((......((.....)).....)))..).))..).))).)))))))))((((((.....)))))). ( -41.50) >consensus UGUUGUAUUUGCUAAUGGGAUGGCCCUGAAUGGGCGU___CGUUCGGGGCGUGGCAAUUAAAGCACCUUGUCCAAGUGCUGCGAUUUCGGGCGGCGUGCAAUCAGUUGAAUUGCAAUUGG .((((((((((((...(((((.((((((((((........))))))))))(..(((......((.....)).....)))..).))))).))))).)))))))((((((.....)))))). (-38.62 = -38.66 + 0.04)

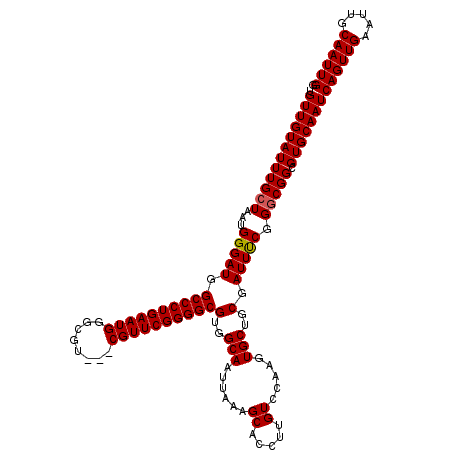
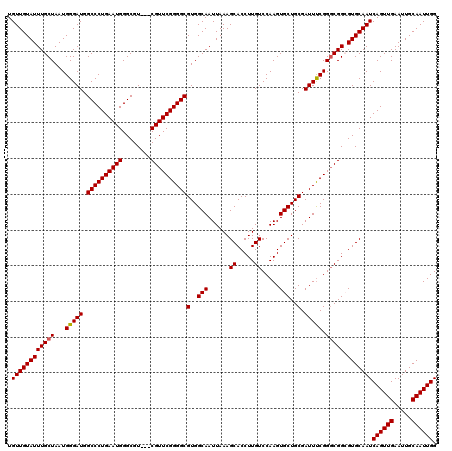
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:55 2006