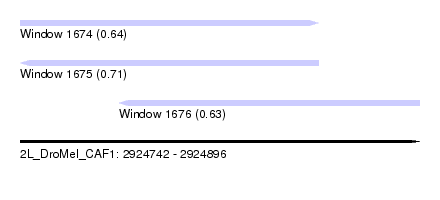
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,924,742 – 2,924,896 |
| Length | 154 |
| Max. P | 0.708558 |

| Location | 2,924,742 – 2,924,857 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.53 |
| Mean single sequence MFE | -21.66 |
| Consensus MFE | -16.62 |
| Energy contribution | -16.78 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2924742 115 + 22407834 CUUUCCUUCGUUUC--UCUGCCUCUUGGCUUACAUUUGGCUUGCAAAUUGUUGCUCUCUAUUUAUCGCUUUCUAGCUGAUUUAAUUAAUUUUAUUUCCACUAGUAG---AUACACAAGCA ..............--...(((....))).........((((((((....))))........((((.....((((..((..(((......)))..))..))))..)---)))...)))). ( -17.70) >DroSec_CAF1 42896 115 + 1 CUUUCCUUCGUUUC--GCUGCCUCUUGGCUUACAUUUGGCUUGCAAAUUGUUGCUCGCUAUUUAUCGCUUUCUAGCUGAUUUAAUUAAUUUUAUUUCCACUAGUAG---AUACACAAGCA ..............--((((((....))).......((((..((((....))))..))))..((((.....((((..((..(((......)))..))..))))..)---)))....))). ( -21.80) >DroSim_CAF1 43772 117 + 1 CUUUCCUUCGUUUCGCGCUGCCUCUUGGCUUACAUUUGGCUUGCAAAUUGUUGCUCGCUAUUUAUCGCUUUCUAGCUGAUUUAAUUAAUUUUAUUUCCACUAGUAG---AUACACAAGCA .........((.(((((..(((....))).......((((..((((....))))..)))).....)))...((((..((..(((......)))..))..))))..)---).))....... ( -22.00) >DroEre_CAF1 43904 118 + 1 CUUUCAUUCGUUUC--GCUGCCUCUUGGCUUACAUUUGGCUUGCAAAUUGUUGCUCGCUAUUUAUCGCUUUCUAGCUGAUUUAAUUUAUUUUAUAUCUACUAGUGGUGUAUAGACAAACA .........((((.--...(((....))).......((((..((((....))))..))))(((((((((..((((..(((.(((......))).)))..)))).)))).))))).)))). ( -24.90) >DroYak_CAF1 43949 115 + 1 CUUUCCUUCGUUUC--GCUGCCUCUUGGCUUACAUUUGGCUUGCAAAUUGUUGCUCGCUAUUUAUCGCUUUCUAGCUGAUUUAAUUAAUUUUAUAUCUACUAGUGG---AUACACAAACA ...(((........--((.(((....))).......((((..((((....))))..))))......))...((((..(((.(((......))).)))..)))).))---).......... ( -21.90) >consensus CUUUCCUUCGUUUC__GCUGCCUCUUGGCUUACAUUUGGCUUGCAAAUUGUUGCUCGCUAUUUAUCGCUUUCUAGCUGAUUUAAUUAAUUUUAUUUCCACUAGUAG___AUACACAAGCA .........((((...((.(((....))).......((((..((((....))))..))))......))...((((..((..(((......)))..))..))))............)))). (-16.62 = -16.78 + 0.16)

| Location | 2,924,742 – 2,924,857 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.53 |
| Mean single sequence MFE | -22.16 |
| Consensus MFE | -18.06 |
| Energy contribution | -18.02 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2924742 115 - 22407834 UGCUUGUGUAU---CUACUAGUGGAAAUAAAAUUAAUUAAAUCAGCUAGAAAGCGAUAAAUAGAGAGCAACAAUUUGCAAGCCAAAUGUAAGCCAAGAGGCAGA--GAAACGAAGGAAAG ..(((.(((.(---((((...(((................(((.(((....)))))).........((((....))))...)))...))..(((....)))..)--)).))).))).... ( -18.90) >DroSec_CAF1 42896 115 - 1 UGCUUGUGUAU---CUACUAGUGGAAAUAAAAUUAAUUAAAUCAGCUAGAAAGCGAUAAAUAGCGAGCAACAAUUUGCAAGCCAAAUGUAAGCCAAGAGGCAGC--GAAACGAAGGAAAG (((((((.(((---(..(((((.((..(((......)))..)).))))).....))))....)))))))....(((((..((.....))..(((....))).))--))).(....).... ( -23.00) >DroSim_CAF1 43772 117 - 1 UGCUUGUGUAU---CUACUAGUGGAAAUAAAAUUAAUUAAAUCAGCUAGAAAGCGAUAAAUAGCGAGCAACAAUUUGCAAGCCAAAUGUAAGCCAAGAGGCAGCGCGAAACGAAGGAAAG (((((((.(((---(..(((((.((..(((......)))..)).))))).....))))....)))))))....(((((..((.....))..(((....)))...))))).(....).... ( -23.10) >DroEre_CAF1 43904 118 - 1 UGUUUGUCUAUACACCACUAGUAGAUAUAAAAUAAAUUAAAUCAGCUAGAAAGCGAUAAAUAGCGAGCAACAAUUUGCAAGCCAAAUGUAAGCCAAGAGGCAGC--GAAACGAAUGAAAG .((((((..........(((((.(((.(((......))).))).)))))...((........((..((((....))))..)).........(((....))).))--...))))))..... ( -22.50) >DroYak_CAF1 43949 115 - 1 UGUUUGUGUAU---CCACUAGUAGAUAUAAAAUUAAUUAAAUCAGCUAGAAAGCGAUAAAUAGCGAGCAACAAUUUGCAAGCCAAAUGUAAGCCAAGAGGCAGC--GAAACGAAGGAAAG (((((((.(((---((.(((((.(((.(((......))).))).)))))...).))))....)))))))....(((((..((.....))..(((....))).))--))).(....).... ( -23.30) >consensus UGCUUGUGUAU___CUACUAGUGGAAAUAAAAUUAAUUAAAUCAGCUAGAAAGCGAUAAAUAGCGAGCAACAAUUUGCAAGCCAAAUGUAAGCCAAGAGGCAGC__GAAACGAAGGAAAG (((((((.(((......(((((.((..(((......)))..)).)))))......)))....))))))).....(((((.......)))))(((....)))................... (-18.06 = -18.02 + -0.04)

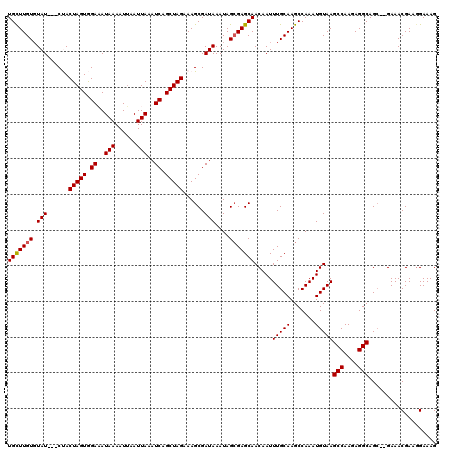
| Location | 2,924,780 – 2,924,896 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -24.31 |
| Consensus MFE | -18.24 |
| Energy contribution | -19.44 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
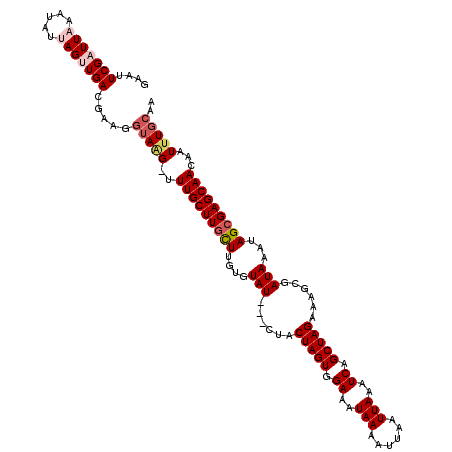
>2L_DroMel_CAF1 2924780 116 - 22407834 GAAUUCGAUUAAAUAUUAGUUGACGAAGU-AAGGUUUGCUUGCUUGUGUAU---CUACUAGUGGAAAUAAAAUUAAUUAAAUCAGCUAGAAAGCGAUAAAUAGAGAGCAACAAUUUGCAA ....(((((((.....)))))))....((-(((..((((((..((((.(((---(..(((((.((..(((......)))..)).))))).....)))).)))).))))))...))))).. ( -21.30) >DroSec_CAF1 42934 117 - 1 GAAUUCGAUUAAAUAUUAGUUGACGAAGGUAGGUUUUGCUUGCUUGUGUAU---CUACUAGUGGAAAUAAAAUUAAUUAAAUCAGCUAGAAAGCGAUAAAUAGCGAGCAACAAUUUGCAA ....(((((((.....))))))).....((((((((((((((((....(((---(..(((((.((..(((......)))..)).))))).....))))...))))))))).))))))).. ( -28.10) >DroSim_CAF1 43812 117 - 1 GAAUUCGAUUAAAUAUUAGUUGACGAAGGUAGGUUUUGCUUGCUUGUGUAU---CUACUAGUGGAAAUAAAAUUAAUUAAAUCAGCUAGAAAGCGAUAAAUAGCGAGCAACAAUUUGCAA ....(((((((.....))))))).....((((((((((((((((....(((---(..(((((.((..(((......)))..)).))))).....))))...))))))))).))))))).. ( -28.10) >DroEre_CAF1 43942 119 - 1 GAAUUCGAUUUAAUAUUAGAUGACAAAGGUAAG-UUUGCUUGUUUGUCUAUACACCACUAGUAGAUAUAAAAUAAAUUAAAUCAGCUAGAAAGCGAUAAAUAGCGAGCAACAAUUUGCAA ....................((.((((.....(-((((((.(((((((.........(((((.(((.(((......))).))).))))).....)))))))))))))).....)))))). ( -21.54) >DroYak_CAF1 43987 116 - 1 GAAUUCGAUUUAAUAUUAGAUGACACAGGUAUG-UUUGCUUGUUUGUGUAU---CCACUAGUAGAUAUAAAAUUAAUUAAAUCAGCUAGAAAGCGAUAAAUAGCGAGCAACAAUUUGCAA .........................(((((.((-((.(((((((....(((---((.(((((.(((.(((......))).))).)))))...).))))...))))))))))))))))... ( -22.50) >consensus GAAUUCGAUUAAAUAUUAGUUGACGAAGGUAAG_UUUGCUUGCUUGUGUAU___CUACUAGUGGAAAUAAAAUUAAUUAAAUCAGCUAGAAAGCGAUAAAUAGCGAGCAACAAUUUGCAA ....(((((((.....))))))).....(((((..(((((((((....(((......(((((.((..(((......)))..)).)))))......)))...)))))))))...))))).. (-18.24 = -19.44 + 1.20)

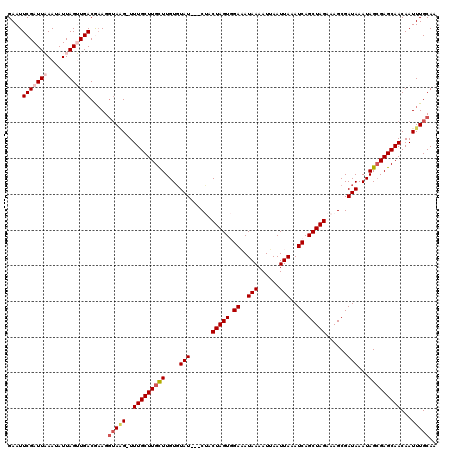
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:51 2006