| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,921,803 – 2,921,921 |
| Length | 118 |
| Max. P | 0.849598 |

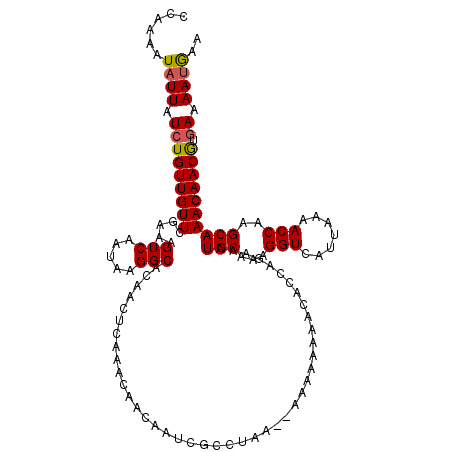
| Location | 2,921,803 – 2,921,921 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.73 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -19.51 |
| Energy contribution | -19.03 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2921803 118 + 22407834 UUUGUUUUUACGUUGUUUGCUUGGUUUUAAUGACCUUUGCAUUUCUGGUGUUUUUUUU--AUAGGUGAUUGUUGUUUGAGCUGUGCCUUAUUGACUUUCGAACAACAGAUAAUAUUUUGG ...........(((((((((..((((.....))))...))).................--.........(((((((((((..((.........)).)))))))))))))))))....... ( -19.70) >DroSec_CAF1 40079 120 + 1 UUCAUUUUCACGUUGUUUGCUUGGUUUUAAUGACCUUUGCAUUUCUGGUGAUUUUUUUUGUUAGGCGAUUGUUGUUUGAGUUGUGCCUUAUUGACUUUCGAACAACAGAUAAUAUUUUGG ..........(((....(((..((((.....))))...)))...((((..(......)..))))))).(((((((((((((((........))))..)))))))))))............ ( -24.10) >DroSim_CAF1 40927 118 + 1 UUCAUUUUCACGUUGUUUGCUUGGUUUUAAUGACCUUUGCAUUUCUGGUGUUUUUUUU--UUAGGUGAUUGUUGUUUGAGUUGUGCCUUAUUGACUUUCGAACAACAGAUAAUAUUUUGG .......((((......(((..((((.....))))...)))...((((..........--))))))))(((((((((((((((........))))..)))))))))))............ ( -22.70) >DroEre_CAF1 40943 115 + 1 UUCAUUUUCAUGUUGUUUGCUUGGUUUUUACGACCUUUGCAUUUCUGCUGUUUAUUU---UUAGGCGAUUGUUGUUCGAGUU--GCCGUAUUGACUUUUGAACAACAGAUAAUAUUUUGG ...(((.((.(((((((((...((((..(((.......(((....))).........---...(((((((........))))--))))))..))))..))))))))))).)))....... ( -25.60) >DroYak_CAF1 41021 115 + 1 UUCAUUUUCAUGUUGUUUGCUUGGUUUUAAUGACCUUUGCAUUUCUGGUGUUUUUUG---UUAGGCGAUUGUUGUUCGAGUU--GCCUUAUUGACUUUUGAACAACAGAUAAUAUUUUGG .........((((((..(((..((((.....))))...)))..((((.(((((...(---((((((((((........))))--))))....)))....))))).))))))))))..... ( -25.80) >consensus UUCAUUUUCACGUUGUUUGCUUGGUUUUAAUGACCUUUGCAUUUCUGGUGUUUUUUUU__UUAGGCGAUUGUUGUUUGAGUUGUGCCUUAUUGACUUUCGAACAACAGAUAAUAUUUUGG ...........(((((((((..((((.....))))...)))............................(((((((((((................)))))))))))))))))....... (-19.51 = -19.03 + -0.48)

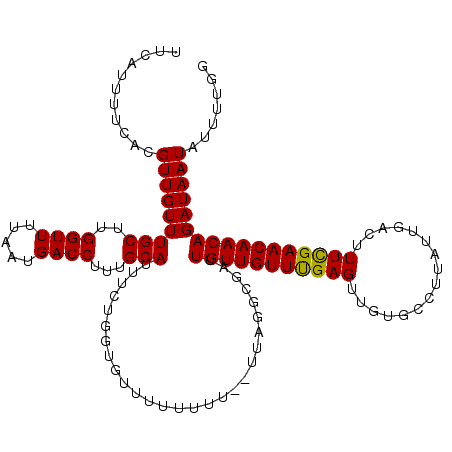
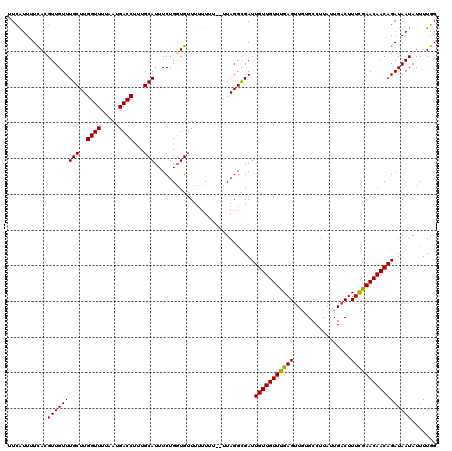
| Location | 2,921,803 – 2,921,921 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.73 |
| Mean single sequence MFE | -15.37 |
| Consensus MFE | -11.28 |
| Energy contribution | -11.28 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2921803 118 - 22407834 CCAAAAUAUUAUCUGUUGUUCGAAAGUCAAUAAGGCACAGCUCAAACAACAAUCACCUAU--AAAAAAAACACCAGAAAUGCAAAGGUCAUUAAAACCAAGCAAACAACGUAAAAACAAA ..............((((((.....(((.....)))...(((..................--...........((....))....(((.......))).))).))))))........... ( -11.80) >DroSec_CAF1 40079 120 - 1 CCAAAAUAUUAUCUGUUGUUCGAAAGUCAAUAAGGCACAACUCAAACAACAAUCGCCUAACAAAAAAAAUCACCAGAAAUGCAAAGGUCAUUAAAACCAAGCAAACAACGUGAAAAUGAA ......((((.(((((((((............((((..................)))).....................(((...(((.......)))..)))))))))).)).)))).. ( -14.87) >DroSim_CAF1 40927 118 - 1 CCAAAAUAUUAUCUGUUGUUCGAAAGUCAAUAAGGCACAACUCAAACAACAAUCACCUAA--AAAAAAAACACCAGAAAUGCAAAGGUCAUUAAAACCAAGCAAACAACGUGAAAAUGAA .............(((((((.((..(((.....))).....)).))))))).((((....--...........((....))....(((.......)))...........))))....... ( -13.40) >DroEre_CAF1 40943 115 - 1 CCAAAAUAUUAUCUGUUGUUCAAAAGUCAAUACGGC--AACUCGAACAACAAUCGCCUAA---AAAUAAACAGCAGAAAUGCAAAGGUCGUAAAAACCAAGCAAACAACAUGAAAAUGAA .............((((((((....(((.....)))--.....))))))))...((....---.........(((....)))...(((.......)))..))......(((....))).. ( -20.70) >DroYak_CAF1 41021 115 - 1 CCAAAAUAUUAUCUGUUGUUCAAAAGUCAAUAAGGC--AACUCGAACAACAAUCGCCUAA---CAAAAAACACCAGAAAUGCAAAGGUCAUUAAAACCAAGCAAACAACAUGAAAAUGAA .............((((((((....(((.....)))--.....))))))))...((....---..........((....))....(((.......)))..))......(((....))).. ( -16.10) >consensus CCAAAAUAUUAUCUGUUGUUCGAAAGUCAAUAAGGCACAACUCAAACAACAAUCGCCUAA__AAAAAAAACACCAGAAAUGCAAAGGUCAUUAAAACCAAGCAAACAACGUGAAAAUGAA ......((((.(((((((((.....(((.....)))...........................................(((...(((.......)))..)))))))))).)).)))).. (-11.28 = -11.28 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:43 2006