
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,915,712 – 2,915,824 |
| Length | 112 |
| Max. P | 0.700449 |

| Location | 2,915,712 – 2,915,824 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.92 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -20.28 |
| Energy contribution | -21.00 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2915712 112 + 22407834 --UCAGCCAGGAUGUAGGAAAUUUUCCGGUUUUACUGUUGAGCCUCCUGCUGCCAGGAAAUCGCUCCUACUGCUG-CGAGCAAAGAACAGA--AAAGCAGCAAAAAAAAACACACU--C- --.((((.((((....(((.....)))(((((.......)))))))))))))..((((......))))..(((((-(...(........).--...))))))..............--.- ( -28.50) >DroSec_CAF1 34158 111 + 1 --UCAGCCAGGAAGUAGGAAAUUUUCCGGUUUUACUGUUGAGCCUCCUGCUGCCAGGAAAUCGCUCCUACUGCUG-CGUUCACAGAACAGA--AAAGCAGCAAAAAA-AACACACU--C- --.......((.(((((((..(((..(((.....)))..)))..))))))).))((((......))))..(((((-(.(((........))--)..)))))).....-........--.- ( -30.40) >DroSim_CAF1 34817 110 + 1 --UCAGUCAGGAAGUAGGAAAUUUUCCGGUUUUACUGUUGAGCCUCCUGCUGCCAGGAAAUCGCUCCUACUGCUG-CGUUCAUAGAACAGA--AAAGCAGCAAAAAA--ACACACU--C- --.......((.(((((((..(((..(((.....)))..)))..))))))).))((((......))))..(((((-(.(((........))--)..)))))).....--.......--.- ( -30.40) >DroEre_CAF1 35023 101 + 1 --UCAGCCAGGAAGUAGGAAAUUUUCCGGUUUUACUGUUGAGCCUCCUGCUGCCAGGAAAUCGCUCCUACUGCUG-C-----------AGG--AAAGCAGAAAAAAA---CACACCAUCA --..((((.(((((........)))))))))...(((((.....((((((.((.((((......))))...)).)-)-----------)))--).))))).......---.......... ( -28.00) >DroYak_CAF1 35151 103 + 1 --UCAGCCAGGAAGUAGGAAAUUUUCCGGUUUUACUGUUGAGCCUCCUGCUGCCAGGAAAUCGCUCCUACUGCUG-CGAGCGCAAAACAGU--AAAGCAGAAAAAAA------------A --.......(((((........))))).(((((((((((..((.(((((....)))))....(((((.......)-.))))))..))))))--))))).........------------. ( -31.80) >DroAna_CAF1 19962 114 + 1 AUUCAGUCAGGAAGUAGGAAAUUUUCCGGCUUUA-UGUUGAGCCUCCCGCUGCCAGGAAAUCGGUGUAGCCCCCGUCGGUCGCCGAGCUUUAGAGUGCAGCAAAAAA-UAUAUGUU---- .........(((....(((.....)))(((((..-....)))))))).((((((..(((.((((((..(((......)))))))))..)))...).)))))......-........---- ( -32.50) >consensus __UCAGCCAGGAAGUAGGAAAUUUUCCGGUUUUACUGUUGAGCCUCCUGCUGCCAGGAAAUCGCUCCUACUGCUG_CGAUCACAGAACAGA__AAAGCAGCAAAAAA__ACACACU__C_ ...(((.(((((....(((.....)))(((((.......)))))))))))))..((((......)))).(((((.....................))))).................... (-20.28 = -21.00 + 0.72)

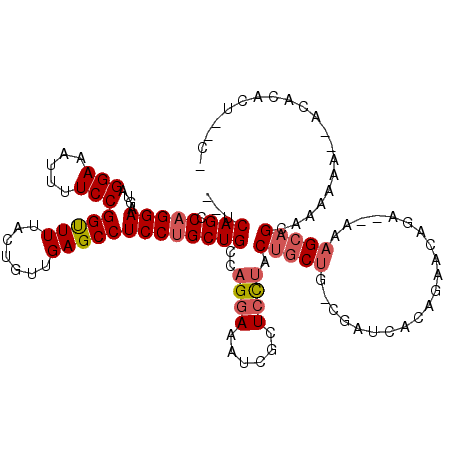
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:34 2006