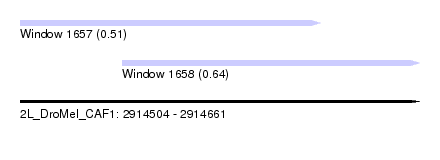
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,914,504 – 2,914,661 |
| Length | 157 |
| Max. P | 0.642382 |

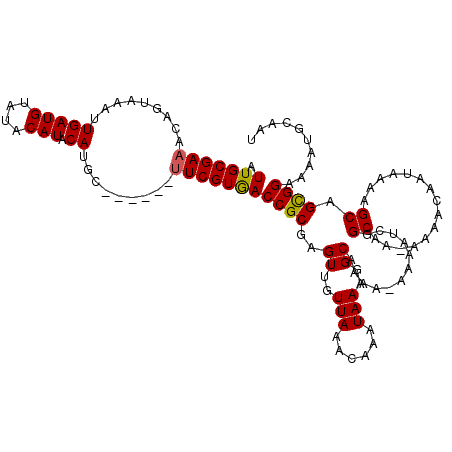
| Location | 2,914,504 – 2,914,622 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.33 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -20.33 |
| Energy contribution | -20.09 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2914504 118 + 22407834 UUUCACUCACAUUUUGUUUGUUGACCUUGUGACUCAGCAUAUUGCGAAACAGUAAAUUGAUGUAUACAUACAUGUACAUGCUUCGUGACCGCGAGUUGUUAAACAAAUAAAGGCAAAA-- ...........(((((((((((......(..((((.((...(..((((.(((((...(((((....))).))..))).)).))))..)..))))))..)..)))))))))))......-- ( -27.40) >DroSec_CAF1 32981 113 + 1 GUUCACUCACAUUUUGUUUGUUGACCUUGUGACUCAGCAUAUUGCGAAACAGUAAAUUGAUGUAUACAUACAUGU------UUCGUAACCGCAAGUUGUUAAACAAAUAAAGGCAACA-G ............(((((((((((((((((((..........(((((((((((((..............))).)))------))))))).))))))..))).))))))))))(....).-. ( -24.04) >DroSim_CAF1 33599 112 + 1 UUUCACUCACAUUUUGUUUGUUGACCUUGUGACUCAGCAUAUUGCGAAACAGUAAAUUGAUGUAUACAUACAUGC-------UCGUGACCGCGAGUUGUUAAACAAAUAAAGGCAACA-G ..................(((((.(((((((((((((..(((((.....)))))..)))).)).)))..(((.((-------((((....)))))))))..........)))))))))-. ( -24.40) >DroEre_CAF1 33833 113 + 1 UUUCAAUUACAUUUUGUUUGUUGACCUUGUGACUCAGCAUAUUGCGAAAGAGUAAAUUGAUGUAUACAUACAAGC------UUCGUGACCGCGAGUAGUUAAACAAAUAAAGGCAAAC-A ...........((((((((((((((((((((((((.((.....))....))))...((((((....))).)))..------........))))))..))).)))))))))))......-. ( -22.20) >DroYak_CAF1 33944 114 + 1 UUUUAAUUACAUUUUGUUUGUUGACCUUGUGGCUCAGCAUAUUGCGAAACAGUAAAUUGAUGUAUACAUACAAGC------UUCGUGACCGCGAGUAGUUAAGCAAAUAAAAGCAAAAAA ...........(((((((((((((((((((((.(((((...((((......)))).((((((....))).)))))------....))))))))))..))).)))))))))))........ ( -25.60) >consensus UUUCACUCACAUUUUGUUUGUUGACCUUGUGACUCAGCAUAUUGCGAAACAGUAAAUUGAUGUAUACAUACAUGC______UUCGUGACCGCGAGUUGUUAAACAAAUAAAGGCAAAA_A ...........(((((((((((......(..((((.((...(((((((.........(((((....))).)).........)))))))..))))))..)..)))))))))))........ (-20.33 = -20.09 + -0.24)

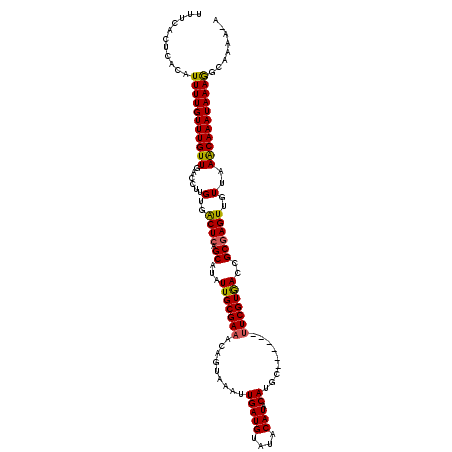
| Location | 2,914,544 – 2,914,661 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.06 |
| Mean single sequence MFE | -18.90 |
| Consensus MFE | -14.29 |
| Energy contribution | -14.17 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2914544 117 + 22407834 AUUGCGAAACAGUAAAUUGAUGUAUACAUACAUGUACAUGCUUCGUGACCGCGAGUUGUUAAACAAAUAAAGGCAAAA--AAAUCGGCAA-AAAACAAUAAAAGCAGUGGAAAAUGCAAU .((((......(((...(((((....))).))..))).((((((((....))))(((....))).......))))...--......))))-............(((.(....).)))... ( -18.50) >DroSec_CAF1 33021 112 + 1 AUUGCGAAACAGUAAAUUGAUGUAUACAUACAUGU------UUCGUAACCGCAAGUUGUUAAACAAAUAAAGGCAACA-GAACUCGGCAA-AAAACAACAAAAGCAGCGGAAAAUGCAAU .(((((((((((((..............))).)))------)))))))((((..((((((...........)))))).-.......((..-............)).)))).......... ( -22.58) >DroSim_CAF1 33639 111 + 1 AUUGCGAAACAGUAAAUUGAUGUAUACAUACAUGC-------UCGUGACCGCGAGUUGUUAAACAAAUAAAGGCAACA-GAACUCGGCAA-AAAACAACAAAAGCAGCGGAAAAUGCAAU ((((((..((((((...(((((....))).)))))-------).))..((((((((((.....).......(....).-.))))).((..-............)).))))....)))))) ( -19.54) >DroEre_CAF1 33873 112 + 1 AUUGCGAAAGAGUAAAUUGAUGUAUACAUACAAGC------UUCGUGACCGCGAGUAGUUAAACAAAUAAAGGCAAAC-AAAAACCGCAA-AAAAUAAUAAAAGCAGCGGAAAAUGCAAU ((((((.....((...((((((....))).)))((------(((((....))))((......)).......)))..))-.....((((..-...............))))....)))))) ( -16.53) >DroYak_CAF1 33984 114 + 1 AUUGCGAAACAGUAAAUUGAUGUAUACAUACAAGC------UUCGUGACCGCGAGUAGUUAAGCAAAUAAAAGCAAAAAAAAAACCGCAACAAAAUAAUAAAAGCAGCGGAAAAUGCAAU .((((..(((......((((((....))).)))((------(.(((....)))))).)))..))))..................((((..................)))).......... ( -17.37) >consensus AUUGCGAAACAGUAAAUUGAUGUAUACAUACAUGC______UUCGUGACCGCGAGUUGUUAAACAAAUAAAGGCAAAA_AAAAUCGGCAA_AAAACAAUAAAAGCAGCGGAAAAUGCAAU .(((((((.........(((((....))).)).........)))))))((((..((..(((......)))..))............((...............)).)))).......... (-14.29 = -14.17 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:33 2006