| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,913,987 – 2,914,211 |
| Length | 224 |
| Max. P | 0.647238 |

| Location | 2,913,987 – 2,914,100 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 84.69 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -23.29 |
| Energy contribution | -23.35 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2913987 113 - 22407834 ACGCGGCGUAUGAGCAAUUUGCUCGCUCUCAAAGCGAAAAUGCGAGCACAUUUCGAAAUGCCAAGCGGAGCAUAUGCUGAAAUCUCUCCUCCAACGCGACACACCCCCCUCCC .(((((((((((.((....(((((((..((.....))....)))))))((((....))))....))....))))))).((....))........))))............... ( -28.00) >DroSec_CAF1 32491 96 - 1 ACGCGGCGUAUGAGCAAUUUGCUCGCUCUCAAAGCGAAAAUGCGAGCACAUUUCGAAAUGCCAAGAGGAGCAUAUGCUGAAAUCUCUGCCGCAU-----------------CC ..(((((((((..(.(((.(((((((..((.....))....))))))).))).)...))))..((((((((....)))....))))))))))..-----------------.. ( -32.50) >DroSim_CAF1 33105 96 - 1 ACGCGGCGUAUGAGCAAUUUGCCCGCUCUCAAAGCGAAAAUGCGAGCACAUUUCGGAAUGCCAAGCGGAGCAUAUGCUGAAAUCUCUGCCCCAU-----------------CC ..((((.(..(.((((...((((((((..((...(((((.((....))..)))))...))...))))).)))..)))).)...).)))).....-----------------.. ( -25.20) >DroYak_CAF1 33447 92 - 1 ACGCGGCGUAUGAGCAAUUUGCUCGCUCUCAAAGCGAAAAUGUGAGCACAUUUCGAAAUGCCAAGCGGAGCAUAUGCUGAAAUCUGA----AAU-----------------CU ...((((((((((((.....))))(((((.....(((((.(((....))))))))....((...)))))))))))))))........----...-----------------.. ( -26.90) >consensus ACGCGGCGUAUGAGCAAUUUGCUCGCUCUCAAAGCGAAAAUGCGAGCACAUUUCGAAAUGCCAAGCGGAGCAUAUGCUGAAAUCUCUGCCCCAU_________________CC ...(((((((((..(....(((((((..((.....))....)))))))((((....)))).......)..))))))))).................................. (-23.29 = -23.35 + 0.06)

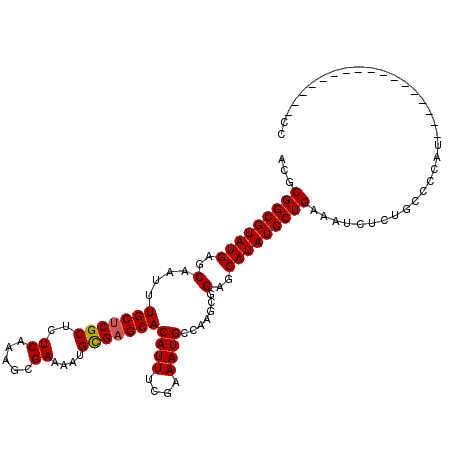
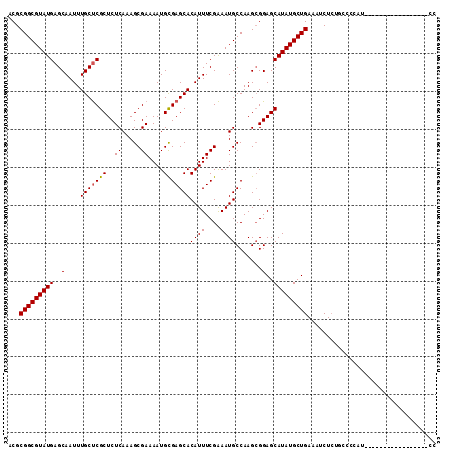
| Location | 2,914,100 – 2,914,211 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.89 |
| Mean single sequence MFE | -37.76 |
| Consensus MFE | -27.63 |
| Energy contribution | -27.51 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2914100 111 + 22407834 UGGGCGCCAGAUGCGGGCGCCAAAGGACGACGACGAGGACGACGACGGUGAAGAAA-----GAACGUC----AGCUCAUAAUUACGGUUAUAUGAGCCGUGAUUAUUCGCUUUUGCCCCU ..((((((.......))))))..(((.((....)).((.(((.((((.........-----...))))----(((..((((((((((((.....))))))))))))..))).)))))))) ( -37.40) >DroSec_CAF1 32587 112 + 1 UGGGCGCCAGAUGCGGGCGCCAAAGGACGACGACG---ACGACGACGGUGAAGAAA-----GCACGUCAGUCAGCUCAUAAUUAUGGUUAUAUGAGCCGUGAUUAUUCGACUUUGCCCCU ..((((((.......))))))...((.(((.(.((---(.(((((((((.......-----)).)))).))).....((((((((((((.....))))))))))))))).).)))))... ( -38.00) >DroSim_CAF1 33201 112 + 1 UGGGCGCCAGAUGCGGGUGCCAAAGGACGACGACG---ACGACGACGGUGAAGAAA-----GCACGUCAGUCAGCUCAUAAUUAUGGUUAUAUGAGCCGUGAUUAUUCGCUUUUGCCCCU ..((((((.......))))))...((.(((.....---..(((((((((.......-----)).)))).)))(((..((((((((((((.....))))))))))))..))).)))))... ( -36.60) >DroEre_CAF1 33433 99 + 1 UGGGCGCGAGAUGCGGGCGAAAAAGGACGCCG------------ACGGUGAAGAAG-----GCACGUC----GGCUCAUAAUUAUGGUUAUAUGAGCCGUGAUUAUUCGCUUUUGCCCCU .(((.((((((.((((.........((.((((------------(((((.......-----)).))))----)))))((((((((((((.....))))))))))))))))))))))))). ( -42.50) >DroYak_CAF1 33539 107 + 1 UGGGCGCGAGAUGCGGGCGAAAAAGGACGACGACG---------ACGAUGAAGAAGAAAGCGCACGUC----AGCUCAUAAUUAUGGUUAUAUGAGCCGUGAUUAUUCGCUUUUGCCCCU .(((.((((((.((((((......(.....)...(---------(((.((............))))))----.))))((((((((((((.....))))))))))))..))))))))))). ( -34.30) >consensus UGGGCGCCAGAUGCGGGCGCCAAAGGACGACGACG___ACGACGACGGUGAAGAAA_____GCACGUC____AGCUCAUAAUUAUGGUUAUAUGAGCCGUGAUUAUUCGCUUUUGCCCCU .(((.((.(((.((((((.......((((...................................)))).....))))((((((((((((.....))))))))))))..)).)))))))). (-27.63 = -27.51 + -0.12)

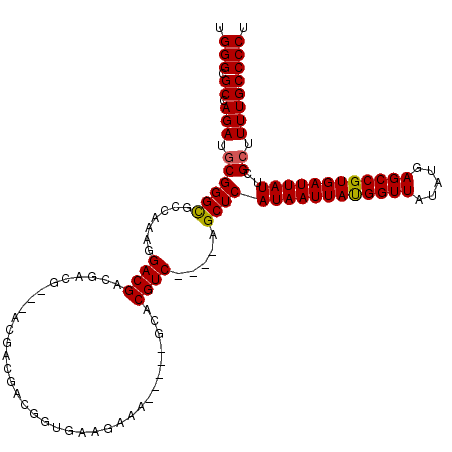
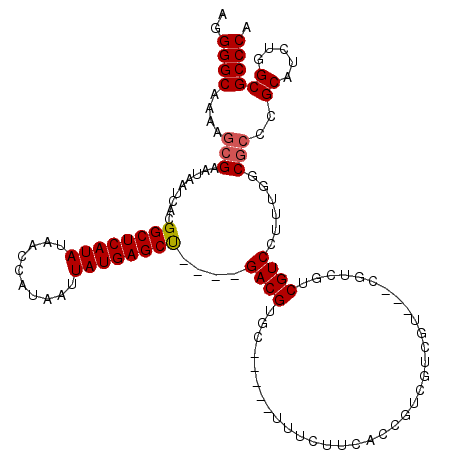
| Location | 2,914,100 – 2,914,211 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.89 |
| Mean single sequence MFE | -34.74 |
| Consensus MFE | -22.53 |
| Energy contribution | -22.77 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2914100 111 - 22407834 AGGGGCAAAAGCGAAUAAUCACGGCUCAUAUAACCGUAAUUAUGAGCU----GACGUUC-----UUUCUUCACCGUCGUCGUCCUCGUCGUCGUCCUUUGGCGCCCGCAUCUGGCGCCCA ((((((......((((.....(((((((((..........))))))))----)..))))-----..........(.((.((....)).)).))))))).((((((.......)))))).. ( -34.90) >DroSec_CAF1 32587 112 - 1 AGGGGCAAAGUCGAAUAAUCACGGCUCAUAUAACCAUAAUUAUGAGCUGACUGACGUGC-----UUUCUUCACCGUCGUCGU---CGUCGUCGUCCUUUGGCGCCCGCAUCUGGCGCCCA ((((((...(.(((........((((((((..........))))))))(((.((((((.-----......)).))))))).)---)).)...)))))).((((((.......)))))).. ( -38.20) >DroSim_CAF1 33201 112 - 1 AGGGGCAAAAGCGAAUAAUCACGGCUCAUAUAACCAUAAUUAUGAGCUGACUGACGUGC-----UUUCUUCACCGUCGUCGU---CGUCGUCGUCCUUUGGCACCCGCAUCUGGCGCCCA .(((((....(((........(((((((((..........)))))))))......((((-----(.....(..((.((....---)).))..)......))))).))).....)).))). ( -33.70) >DroEre_CAF1 33433 99 - 1 AGGGGCAAAAGCGAAUAAUCACGGCUCAUAUAACCAUAAUUAUGAGCC----GACGUGC-----CUUCUUCACCGU------------CGGCGUCCUUUUUCGCCCGCAUCUCGCGCCCA ..((((....((((.......(((((((((..........))))))))----)......-----..........((------------.((((........)))).))...)))))))). ( -34.80) >DroYak_CAF1 33539 107 - 1 AGGGGCAAAAGCGAAUAAUCACGGCUCAUAUAACCAUAAUUAUGAGCU----GACGUGCGCUUUCUUCUUCAUCGU---------CGUCGUCGUCCUUUUUCGCCCGCAUCUCGCGCCCA ..((((....(((((.((..((((((((((..........))))))))----((((.((((.............).---------)))))))))..)).)))))..((.....)))))). ( -32.12) >consensus AGGGGCAAAAGCGAAUAAUCACGGCUCAUAUAACCAUAAUUAUGAGCU____GACGUGC_____UUUCUUCACCGUCGUCGU___CGUCGUCGUCCUUUGGCGCCCGCAUCUGGCGCCCA ..((((....(((.........((((((((..........))))))))....((((...................................))))......)))..((.....)))))). (-22.53 = -22.77 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:31 2006