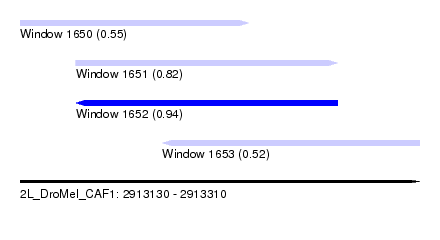
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,913,130 – 2,913,310 |
| Length | 180 |
| Max. P | 0.940207 |

| Location | 2,913,130 – 2,913,233 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.92 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -24.67 |
| Energy contribution | -24.63 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2913130 103 + 22407834 GUUCAAAUGUCAUCGC---------------CGUCGUAGCUAGCCAA-AUUGCCGGGCAUGCCCACAAAAUACCUUCAUGGAACCAAGCCAUGGCUACGGCAUUUCCGAGGACACGG-CU ..............((---------------(((((((((.......-......(((....)))............(((((.......)))))))))))))...((....))...))-). ( -30.30) >DroSec_CAF1 31633 103 + 1 GUUCAAAUGUCAUCGG---------------CGUCGUAGCUAGCCAA-AUUGCCGGGCAUGCCCACAAAAUGCCUUCAAGGAAGCAAGCCAUGGCUACGGCAUUUCCGAGGACACGG-CU .......((((.((((---------------.((((((((((.....-.((((.(((....)))........((.....))..))))....))))))))))....)))).))))...-.. ( -35.40) >DroSim_CAF1 32238 103 + 1 GUUCAAAUGUCAUCGG---------------CGUCGUAGCUAGCCAA-AUUGCCGGGCAUGCCCACAAAAUGCCUUCAAGGAAGCAAGCCAUGGCUACGGCAUUUCCGAGGACACGG-AU ..((...((((.((((---------------.((((((((((.....-.((((.(((....)))........((.....))..))))....))))))))))....)))).))))..)-). ( -35.90) >DroEre_CAF1 32464 119 + 1 GUUCAAAUGUCAUCGUAGUCGUAGUCGUAGUCGUCGUAGCCAGCCAAAAUGGCCGGGCAUGCCCACAAAAUGCCUUCAAGGAACCAAGCCAUGGCUACGGAAUUUCCGAGGACACGG-CU ......(((.(......).)))((((((.(((.((((((((((((.....))).((((((.........))))))....((.......)).)))))))((.....)))).)))))))-)) ( -39.40) >DroYak_CAF1 32617 101 + 1 GUUCAAAUGUCAUCGUG------------------GUAGCCAACCAA-AUUGCCGGGCAUGCCCACAAAAUGCCUUCAAGGAACCAAGCCAUGGCUACGGAAUUUCCGAGGACACGGACU ((((...((((.((...------------------(((((((.....-.(((..((((((.........)))))).)))((.......)).)))))))((.....)))).)))).)))). ( -30.30) >consensus GUUCAAAUGUCAUCGG_______________CGUCGUAGCUAGCCAA_AUUGCCGGGCAUGCCCACAAAAUGCCUUCAAGGAACCAAGCCAUGGCUACGGCAUUUCCGAGGACACGG_CU .......((((.((.....................(((((((..........((((((((.........))))))....))..........)))))))((.....)))).))))...... (-24.67 = -24.63 + -0.04)

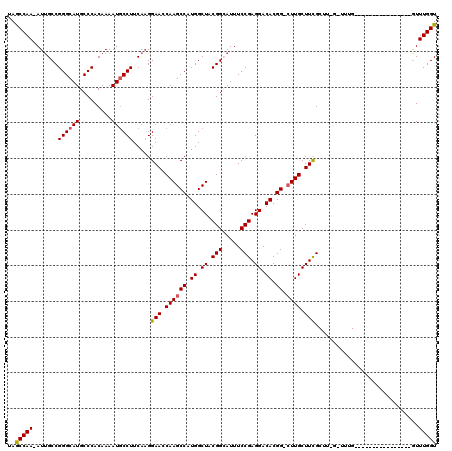
| Location | 2,913,155 – 2,913,273 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -22.36 |
| Energy contribution | -22.36 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2913155 118 + 22407834 UAGCCAA-AUUGCCGGGCAUGCCCACAAAAUACCUUCAUGGAACCAAGCCAUGGCUACGGCAUUUCCGAGGACACGG-CUUGCUUCGCUUCGCUUUGGUUUUGGUUUGCUUCGGUUUGGU ..(((((-...((((((....)))................(((.(((((((..((((.(((.....(((((.((...-..)))))))....))).))))..))))))).))))))))))) ( -38.00) >DroSec_CAF1 31658 102 + 1 UAGCCAA-AUUGCCGGGCAUGCCCACAAAAUGCCUUCAAGGAAGCAAGCCAUGGCUACGGCAUUUCCGAGGACACGG-CUUGCUUCGCUUUGGUUUG----------------GUUUGGU ..(((((-((((((((((((.........))))))..(((((((((((((.((.((.(((.....))).)).)).))-)))))))).))).)))..)----------------))))))) ( -42.60) >DroSim_CAF1 32263 113 + 1 UAGCCAA-AUUGCCGGGCAUGCCCACAAAAUGCCUUCAAGGAAGCAAGCCAUGGCUACGGCAUUUCCGAGGACACGG-AUUGCUUCGCUUUGGUUUGGUUCAG-----UUUUGGUUUGGU ..(((((-...(((((((((.........)))))).....(((.(((((((.(((...((((..((((......)))-).))))..))).))))))).)))..-----....)))))))) ( -41.20) >DroEre_CAF1 32504 93 + 1 CAGCCAAAAUGGCCGGGCAUGCCCACAAAAUGCCUUCAAGGAACCAAGCCAUGGCUACGGAAUUUCCGAGGACACGG-CUUGCUUUG--------------------------CUUUGGU ..(((.....))).((((((.........))))))((((((...((((((.((.((.(((.....))).)).)).))-)))).....--------------------------)))))). ( -31.70) >DroYak_CAF1 32639 93 + 1 CAACCAA-AUUGCCGGGCAUGCCCACAAAAUGCCUUCAAGGAACCAAGCCAUGGCUACGGAAUUUCCGAGGACACGGACUUGCUUUG--------------------------GUUUGGU ..(((((-((....((((((.........)))))).(((((...((((((.((.((.(((.....))).)).)).)).)))))))))--------------------------))))))) ( -28.60) >consensus UAGCCAA_AUUGCCGGGCAUGCCCACAAAAUGCCUUCAAGGAACCAAGCCAUGGCUACGGCAUUUCCGAGGACACGG_CUUGCUUCGCUU_G_UUUG________________GUUUGGU ..(((((.......((((((.........)))))).....(((.((((((.((.((.(((.....))).)).)).)).)))).))).............................))))) (-22.36 = -22.36 + -0.00)

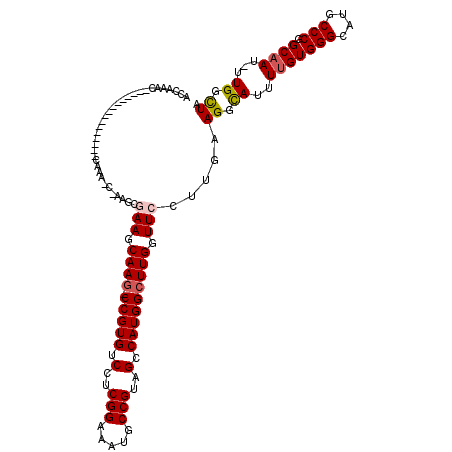
| Location | 2,913,155 – 2,913,273 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -25.24 |
| Energy contribution | -25.72 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2913155 118 - 22407834 ACCAAACCGAAGCAAACCAAAACCAAAGCGAAGCGAAGCAAG-CCGUGUCCUCGGAAAUGCCGUAGCCAUGGCUUGGUUCCAUGAAGGUAUUUUGUGGGCAUGCCCGGCAAU-UUGGCUA .((((((((..(((..(((..(((...((...))(((.((((-(((((.(..(((.....)))..).))))))))).)))......)))......)))...))).)))...)-))))... ( -37.90) >DroSec_CAF1 31658 102 - 1 ACCAAAC----------------CAAACCAAAGCGAAGCAAG-CCGUGUCCUCGGAAAUGCCGUAGCCAUGGCUUGCUUCCUUGAAGGCAUUUUGUGGGCAUGCCCGGCAAU-UUGGCUA .......----------------.......(((.((((((((-(((((.(..(((.....)))..).))))))))))))))))..((.((..(((((((....))).)))).-.)).)). ( -42.40) >DroSim_CAF1 32263 113 - 1 ACCAAACCAAAA-----CUGAACCAAACCAAAGCGAAGCAAU-CCGUGUCCUCGGAAAUGCCGUAGCCAUGGCUUGCUUCCUUGAAGGCAUUUUGUGGGCAUGCCCGGCAAU-UUGGCUA ............-----.............(((.(((((((.-(((((.(..(((.....)))..).))))).))))))))))..((.((..(((((((....))).)))).-.)).)). ( -35.80) >DroEre_CAF1 32504 93 - 1 ACCAAAG--------------------------CAAAGCAAG-CCGUGUCCUCGGAAAUUCCGUAGCCAUGGCUUGGUUCCUUGAAGGCAUUUUGUGGGCAUGCCCGGCCAUUUUGGCUG ......(--------------------------(((((((((-(((((.(..(((.....)))..).)))))))))...((.....))..))))))(((....)))((((.....)))). ( -33.90) >DroYak_CAF1 32639 93 - 1 ACCAAAC--------------------------CAAAGCAAGUCCGUGUCCUCGGAAAUUCCGUAGCCAUGGCUUGGUUCCUUGAAGGCAUUUUGUGGGCAUGCCCGGCAAU-UUGGUUG ....(((--------------------------((((.((((.(((((.(..(((.....)))..).)))))))))................(((((((....))).)))))-)))))). ( -30.00) >consensus ACCAAAC________________CAAA_C_AAGCGAAGCAAG_CCGUGUCCUCGGAAAUGCCGUAGCCAUGGCUUGGUUCCUUGAAGGCAUUUUGUGGGCAUGCCCGGCAAU_UUGGCUA ..................................(((.((((.(((((.(..(((.....)))..).))))))))).))).....((.((..(((((((....))).))))...)).)). (-25.24 = -25.72 + 0.48)

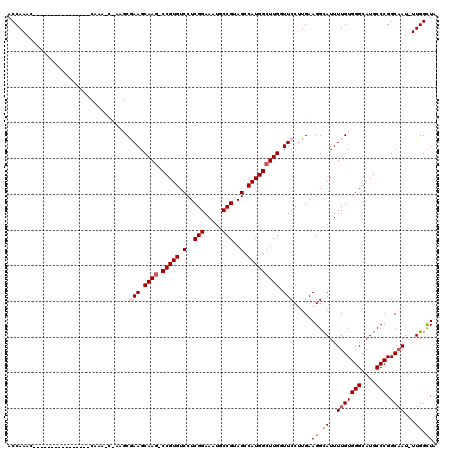
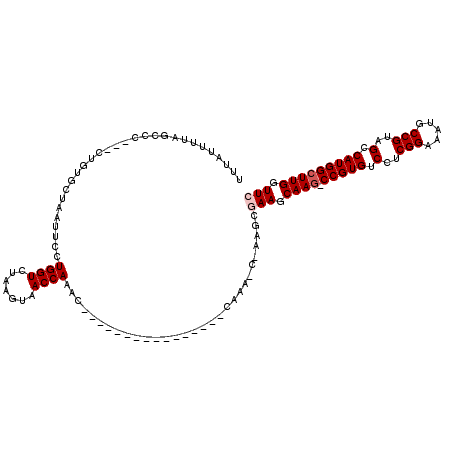
| Location | 2,913,194 – 2,913,310 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -17.50 |
| Energy contribution | -18.10 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2913194 116 - 22407834 UUUAUUUUAGCCC---CUGUGCUAAUUCCUGGUCUAAGUAACCAAACCGAAGCAAACCAAAACCAAAGCGAAGCGAAGCAAG-CCGUGUCCUCGGAAAUGCCGUAGCCAUGGCUUGGUUC ......(((((..---....)))))(((.((((.......)))).......((..............)))))..(((.((((-(((((.(..(((.....)))..).))))))))).))) ( -28.54) >DroSec_CAF1 31697 100 - 1 UUUAUUUUAGCCC---CUGUGCUAAUUCCUGGUCUAAGUAACCAAAC----------------CAAACCAAAGCGAAGCAAG-CCGUGUCCUCGGAAAUGCCGUAGCCAUGGCUUGCUUC ......(((((..---....)))))....((((.......))))...----------------...........((((((((-(((((.(..(((.....)))..).))))))))))))) ( -32.70) >DroSim_CAF1 32302 111 - 1 UUUAUUUUAGUCC---CUGUGCUAAUUCCUGGUCUAAGUAACCAAACCAAAA-----CUGAACCAAACCAAAGCGAAGCAAU-CCGUGUCCUCGGAAAUGCCGUAGCCAUGGCUUGCUUC .............---....(((......((((...(((............)-----))..))))......)))(((((((.-(((((.(..(((.....)))..).))))).))))))) ( -26.60) >DroEre_CAF1 32544 90 - 1 UUGAUUAUGCCCC---CUGUGCUAAUUCCUGGUCUAAGUAACCAAAG--------------------------CAAAGCAAG-CCGUGUCCUCGGAAAUUCCGUAGCCAUGGCUUGGUUC .............---((.((((......((((.......)))).))--------------------------)).))((((-(((((.(..(((.....)))..).))))))))).... ( -24.70) >DroYak_CAF1 32678 94 - 1 UUUAUAAUUCCCCCUCCUGUGCUAAUUCCUGGUCUAAGUAACCAAAC--------------------------CAAAGCAAGUCCGUGUCCUCGGAAAUUCCGUAGCCAUGGCUUGGUUC ............((......(((......((((.......))))...--------------------------...)))(((.(((((.(..(((.....)))..).))))))))))... ( -19.10) >consensus UUUAUUUUAGCCC___CUGUGCUAAUUCCUGGUCUAAGUAACCAAAC________________CAAA_C_AAGCGAAGCAAG_CCGUGUCCUCGGAAAUGCCGUAGCCAUGGCUUGGUUC .............................((((.......))))..............................(((.((((.(((((.(..(((.....)))..).))))))))).))) (-17.50 = -18.10 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:29 2006