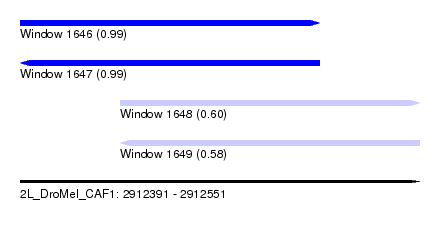
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,912,391 – 2,912,551 |
| Length | 160 |
| Max. P | 0.993289 |

| Location | 2,912,391 – 2,912,511 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -21.80 |
| Energy contribution | -22.24 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2912391 120 + 22407834 UUUCAUUUGUAAUCGUUUUUGCAAUUGUAUUCAAUUGUUGUGACAUUUUAAUUGCUCGCAAUGUAAGCGCUGUAAGCAUAAAUUACGUAAUUAAUUUAUGCUGUACAACUUUUAAUCUUU ..............((.((((((((((....)))))(((((((((.......)).)))))))))))).))(((((((((((((((......))))))))))).))))............. ( -25.60) >DroSec_CAF1 30879 120 + 1 UUUCAGUUGUAAUCAUUUUUGCAAUUGUAUUCAAUUGUUGUGACAUUUUAAUUGAUCGCAAUGUAAGCGCUGUAAGCAUAAAUUACGUUAUUAAUUUAUGCUGUACGACUUUUAAGCUUU ....(((((((......((((((.((((..(((((((...........)))))))..)))))))))).......(((((((((((......))))))))))).))))))).......... ( -28.00) >DroSim_CAF1 31475 120 + 1 UUUCAGUUGUAAUCAUUUUUGCAAUUGUAUUCAAUUGUUGUGACAUUUUAAUUGCUCGCAAUGUAAGCGCUGUAAGCAUAAAUUACGUUAUUAAUUUAUGCUGUACGACUUUUAAGCUUU ....(((((((.((((....(((((((....))))))).)))).........(((.(((.......)))..)))(((((((((((......))))))))))).))))))).......... ( -28.60) >DroEre_CAF1 31666 120 + 1 UUUCAUUUGUAAUCAUUUUUGCAAUUGUAUUCAAUUGUUGUGACAUUUUAAUUGCUCGCAAUGUAAGCGCUGUAAGCAUAAAUUACGUAAUUAUUUUAUCCUCUACGACUUUUAAUCGUU ........((((((((....(((((((....))))))).)))).........((((..((.((....)).))..))))....))))..................((((.......)))). ( -19.10) >DroYak_CAF1 31853 120 + 1 UUUCAUUUGUAAUCAUUUUUGCAAUUGUAUUCAAUUGUUGUGACAUUUUAAUUGCUCGCAAUGUAAGCGCUGCAAGCAUAAAUUACGUAAUUAAUUUAUGCUGUACGGCUUUUAAUCUUU ...((.((((((......)))))).)).........(((((((((.......)).))))))).((((.((((..(((((((((((......)))))))))))...)))).))))...... ( -28.60) >consensus UUUCAUUUGUAAUCAUUUUUGCAAUUGUAUUCAAUUGUUGUGACAUUUUAAUUGCUCGCAAUGUAAGCGCUGUAAGCAUAAAUUACGUAAUUAAUUUAUGCUGUACGACUUUUAAUCUUU ...((.((((((......)))))).)).........(((((((((.......)).))))))).((((.(.(((((((((((((((......))))))))))).)))).).))))...... (-21.80 = -22.24 + 0.44)

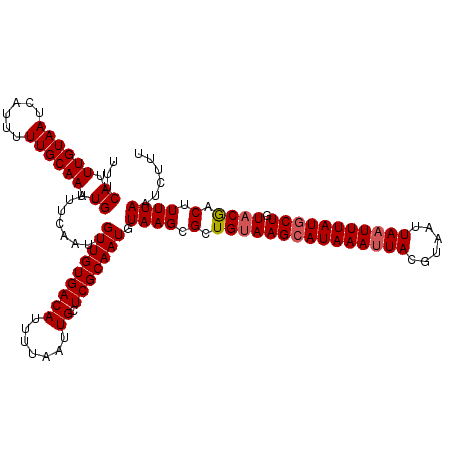
| Location | 2,912,391 – 2,912,511 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -22.26 |
| Consensus MFE | -18.38 |
| Energy contribution | -18.46 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2912391 120 - 22407834 AAAGAUUAAAAGUUGUACAGCAUAAAUUAAUUACGUAAUUUAUGCUUACAGCGCUUACAUUGCGAGCAAUUAAAAUGUCACAACAAUUGAAUACAAUUGCAAAAACGAUUACAAAUGAAA ...(((.....((((((.(((((((((((......)))))))))))))))))((((.......)))).........)))....((((((....))))))..................... ( -25.30) >DroSec_CAF1 30879 120 - 1 AAAGCUUAAAAGUCGUACAGCAUAAAUUAAUAACGUAAUUUAUGCUUACAGCGCUUACAUUGCGAUCAAUUAAAAUGUCACAACAAUUGAAUACAAUUGCAAAAAUGAUUACAACUGAAA .((((......((.(((.(((((((((((......)))))))))))))).)))))).(((((..((((.......((...)).((((((....))))))......))))..))).))... ( -21.50) >DroSim_CAF1 31475 120 - 1 AAAGCUUAAAAGUCGUACAGCAUAAAUUAAUAACGUAAUUUAUGCUUACAGCGCUUACAUUGCGAGCAAUUAAAAUGUCACAACAAUUGAAUACAAUUGCAAAAAUGAUUACAACUGAAA ...((((....((.(((.(((((((((((......)))))))))))))).))((.......)))))).........((((...((((((....))))))......))))........... ( -23.70) >DroEre_CAF1 31666 120 - 1 AACGAUUAAAAGUCGUAGAGGAUAAAAUAAUUACGUAAUUUAUGCUUACAGCGCUUACAUUGCGAGCAAUUAAAAUGUCACAACAAUUGAAUACAAUUGCAAAAAUGAUUACAAAUGAAA .(((((.....)))))...........((((((((((((.((.((.......)).)).)))))).((((((......(((.......)))....)))))).....))))))......... ( -17.70) >DroYak_CAF1 31853 120 - 1 AAAGAUUAAAAGCCGUACAGCAUAAAUUAAUUACGUAAUUUAUGCUUGCAGCGCUUACAUUGCGAGCAAUUAAAAUGUCACAACAAUUGAAUACAAUUGCAAAAAUGAUUACAAAUGAAA ...(((.....((.(((.(((((((((((......)))))))))))))).))((((.......)))).........)))....((((((....))))))..................... ( -23.10) >consensus AAAGAUUAAAAGUCGUACAGCAUAAAUUAAUUACGUAAUUUAUGCUUACAGCGCUUACAUUGCGAGCAAUUAAAAUGUCACAACAAUUGAAUACAAUUGCAAAAAUGAUUACAAAUGAAA ...........((.(((.(((((((((((......)))))))))))))).))....(((((..((....))..))))).....((((((....))))))..................... (-18.38 = -18.46 + 0.08)

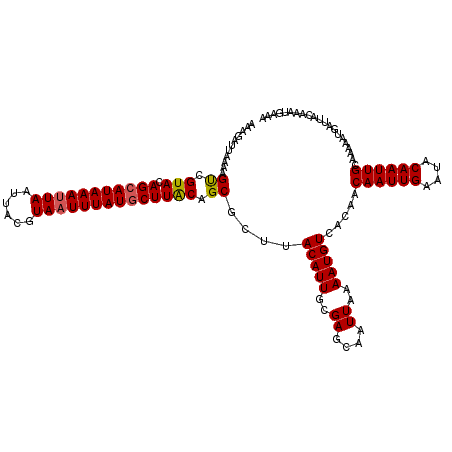
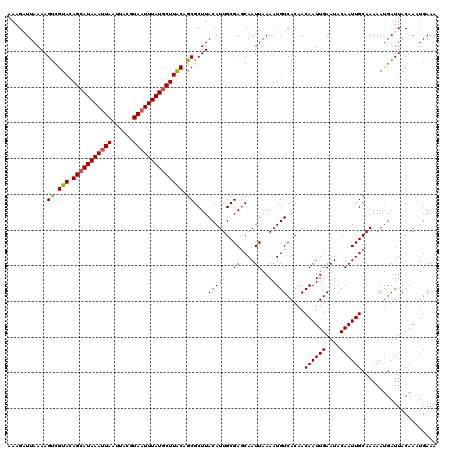
| Location | 2,912,431 – 2,912,551 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.94 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2912431 120 + 22407834 UGACAUUUUAAUUGCUCGCAAUGUAAGCGCUGUAAGCAUAAAUUACGUAAUUAAUUUAUGCUGUACAACUUUUAAUCUUUAUUUCACAGUAAAUAACUUGCAUCGGUCAUUUAAGUGGAA ...(((((.(((.((.((..(((((((...(((((((((((((((......))))))))))).))))..........(((((......)))))...))))))))))).))).)))))... ( -25.30) >DroSec_CAF1 30919 117 + 1 UGACAUUUUAAUUGAUCGCAAUGUAAGCGCUGUAAGCAUAAAUUACGUUAUUAAUUUAUGCUGUACGACUUUUAAGCUUUAUUUCACAGUAA---ACUUGCAUCGGUCAUUUAAGUGGAA ...(((((....((((((..(((((((.(((((.(((((((((((......)))))))))))....(.((....)))........)))))..---.)))))))))))))...)))))... ( -30.00) >DroSim_CAF1 31515 120 + 1 UGACAUUUUAAUUGCUCGCAAUGUAAGCGCUGUAAGCAUAAAUUACGUUAUUAAUUUAUGCUGUACGACUUUUAAGCUUUAUUUCACAGUAAAUAACUUGCAUCGGUCAUUUAAGUGGAA ...(((((.(((.((.((..(((((((...(((((((((((((((......))))))))))).))))..........(((((......)))))...))))))))))).))).)))))... ( -24.60) >DroEre_CAF1 31706 120 + 1 UGACAUUUUAAUUGCUCGCAAUGUAAGCGCUGUAAGCAUAAAUUACGUAAUUAUUUUAUCCUCUACGACUUUUAAUCGUUAUUCCAUAGUAAAUAACUUGCAUUGGUCAUUUAACUGAAA ((((........((((..((.((....)).))..))))........((((((((((........((((.......))))...........)))))).))))....))))........... ( -17.01) >DroYak_CAF1 31893 120 + 1 UGACAUUUUAAUUGCUCGCAAUGUAAGCGCUGCAAGCAUAAAUUACGUAAUUAAUUUAUGCUGUACGGCUUUUAAUCUUUAUUAUACAGUAAAUAACUUGCAUCGGUCAUUUAACUGGAA ((((......((((....))))(((((.((((..(((((((((((......)))))))))))...))))........((((((....))))))...)))))....))))........... ( -26.20) >consensus UGACAUUUUAAUUGCUCGCAAUGUAAGCGCUGUAAGCAUAAAUUACGUAAUUAAUUUAUGCUGUACGACUUUUAAUCUUUAUUUCACAGUAAAUAACUUGCAUCGGUCAUUUAAGUGGAA ...(((((....((.(((..(((((((...(((((((((((((((......))))))))))).))))...........((((......))))....)))))))))).))...)))))... (-19.26 = -19.94 + 0.68)

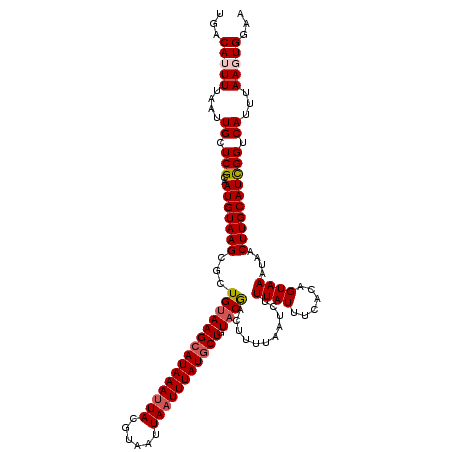
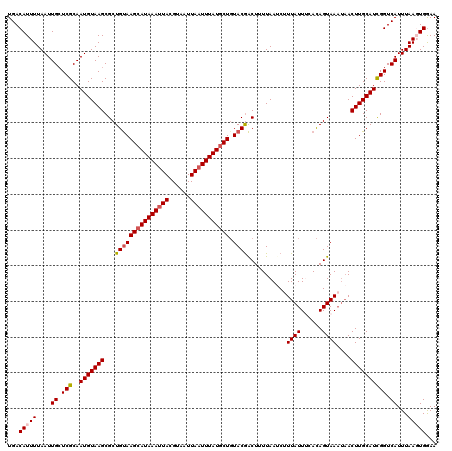
| Location | 2,912,431 – 2,912,551 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -18.40 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2912431 120 - 22407834 UUCCACUUAAAUGACCGAUGCAAGUUAUUUACUGUGAAAUAAAGAUUAAAAGUUGUACAGCAUAAAUUAAUUACGUAAUUUAUGCUUACAGCGCUUACAUUGCGAGCAAUUAAAAUGUCA ...(((.((((((((........))))))))..))).......(((.....((((((.(((((((((((......)))))))))))))))))((((.......)))).........))). ( -29.50) >DroSec_CAF1 30919 117 - 1 UUCCACUUAAAUGACCGAUGCAAGU---UUACUGUGAAAUAAAGCUUAAAAGUCGUACAGCAUAAAUUAAUAACGUAAUUUAUGCUUACAGCGCUUACAUUGCGAUCAAUUAAAAUGUCA ......((((.(((((((((.((((---...((((((..((..(((....)))..))..((((((((((......)))))))))))))))).)))).))))).).))).))))....... ( -24.70) >DroSim_CAF1 31515 120 - 1 UUCCACUUAAAUGACCGAUGCAAGUUAUUUACUGUGAAAUAAAGCUUAAAAGUCGUACAGCAUAAAUUAAUAACGUAAUUUAUGCUUACAGCGCUUACAUUGCGAGCAAUUAAAAUGUCA ...(((.((((((((........))))))))..))).......((((....((.(((.(((((((((((......)))))))))))))).))((.......))))))............. ( -27.70) >DroEre_CAF1 31706 120 - 1 UUUCAGUUAAAUGACCAAUGCAAGUUAUUUACUAUGGAAUAACGAUUAAAAGUCGUAGAGGAUAAAAUAAUUACGUAAUUUAUGCUUACAGCGCUUACAUUGCGAGCAAUUAAAAUGUCA (((((..((((((((........))))))))...)))))..(((((.....)))))....((((...(((((.((((((.((.((.......)).)).))))))...)))))...)))). ( -20.50) >DroYak_CAF1 31893 120 - 1 UUCCAGUUAAAUGACCGAUGCAAGUUAUUUACUGUAUAAUAAAGAUUAAAAGCCGUACAGCAUAAAUUAAUUACGUAAUUUAUGCUUGCAGCGCUUACAUUGCGAGCAAUUAAAAUGUCA ...((((.(((((((........))))))))))).........(((.....((.(((.(((((((((((......)))))))))))))).))((((.......)))).........))). ( -27.20) >consensus UUCCACUUAAAUGACCGAUGCAAGUUAUUUACUGUGAAAUAAAGAUUAAAAGUCGUACAGCAUAAAUUAAUUACGUAAUUUAUGCUUACAGCGCUUACAUUGCGAGCAAUUAAAAUGUCA ......((((.((..(((((.(((((((((......)))))..........((.(((.(((((((((((......)))))))))))))).)))))).)))))....)).))))....... (-18.40 = -19.00 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:25 2006