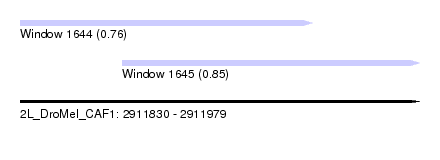
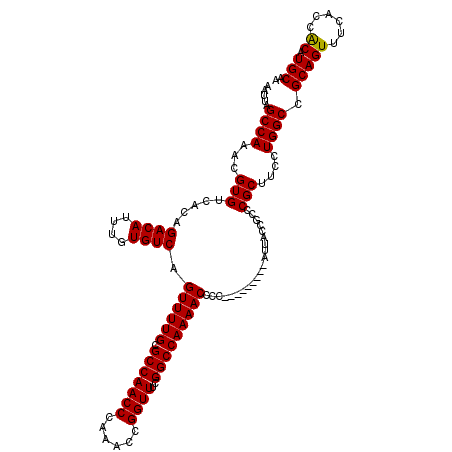
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,911,830 – 2,911,979 |
| Length | 149 |
| Max. P | 0.854070 |

| Location | 2,911,830 – 2,911,939 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.17 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -19.90 |
| Energy contribution | -21.15 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2911830 109 + 22407834 AUGUUUAUCCAAAGCUGUCUAUAUCU--CCGUUUGAUAUAAAUUAGCCAAACGUGUCACAGACAUUUGUGUCCGUUUUGUGCCAACCCAAACCGGUUCGGGCCAAAACCCC--------- .............((((..((((((.--......))))))...)))).....(.(.(((((....))))).))((((((.(((.(((......)))...)))))))))...--------- ( -24.70) >DroSec_CAF1 30321 112 + 1 AUGUUUAUCGAAAGCUGUCCCU--------GUUUGAUAUAAAUUAGCCAAACGUGUCACAGACAUUUGUGUCAGUUUUGUGCCAACCCAAACCGGUUGGGGCCAAAACCCCACUACCUCC ((((....(....)((((..((--------(((((.((.....))..)))))).)..))))))))..(((...(((((((.((((((......)))))).).))))))..)))....... ( -25.70) >DroEre_CAF1 31105 110 + 1 AUGUUUAUCCAAAGCUGUCUAUAUCUCCCUG-UUGAUAUAAAUUAGCCAAACGUGUCACAGACAUUCGUGUCAGUUUUGCGCCAACCCAAACCGGUUUCGGCCAAAACCCC--------- .............((((..((((((......-..))))))...)))).............((((....)))).((((((.(((((((......))))..)))))))))...--------- ( -24.50) >DroYak_CAF1 31334 108 + 1 AUGUUUAUCCAAAGCUGUCUAUAUCU--CUG-UUGAUAUAAAUUAGCCAAACGUGUCACAGACAUAUGUGUCAGUUUUGCGCCAACCCAAACCGGUUUCGGCCAAAACCCC--------- ..((((...((((((((.((((((.(--(((-(.(((((.............)))))))))).))))).).)))))))).(((((((......))))..)))..))))...--------- ( -27.62) >consensus AUGUUUAUCCAAAGCUGUCUAUAUCU__CUG_UUGAUAUAAAUUAGCCAAACGUGUCACAGACAUUUGUGUCAGUUUUGCGCCAACCCAAACCGGUUUCGGCCAAAACCCC_________ .............((((..((((((.........))))))...)))).............((((....)))).((((((.(((((((......))))..)))))))))............ (-19.90 = -21.15 + 1.25)

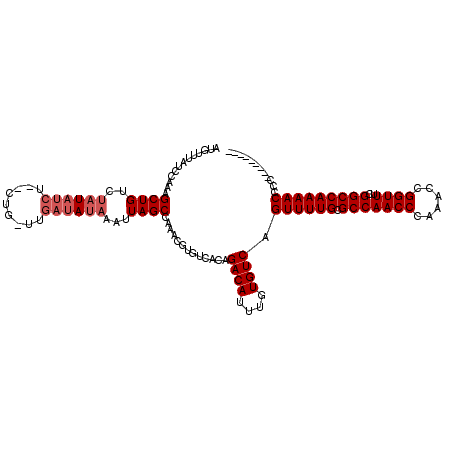
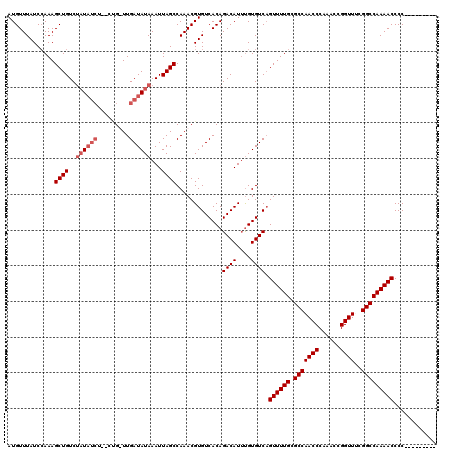
| Location | 2,911,868 – 2,911,979 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -25.76 |
| Energy contribution | -25.57 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2911868 111 + 22407834 AAUUAGCCAAACGUGUCACAGACAUUUGUGUCCGUUUUGUGCCAACCCAAACCGGUUCGGGCCAAAACCCC---------AUUACCGCCCGCUUCCUGGCCGCAGUUUCACCGCAUGCAA .....((.(((((.(.(((((....))))).)))))).))(((((((......))))(((((.........---------......)))))......))).(((((......)).))).. ( -26.46) >DroSec_CAF1 30353 120 + 1 AAUUAGCCAAACGUGUCACAGACAUUUGUGUCAGUUUUGUGCCAACCCAAACCGGUUGGGGCCAAAACCCCACUACCUCCAUUACCUCCCGCUUCCUGGCCGCAGUUUCACCACAUGCAA .....((((....((.(((((....))))).))(((((((.((((((......)))))).).))))))............................)))).(((((......)).))).. ( -28.30) >DroEre_CAF1 31144 111 + 1 AAUUAGCCAAACGUGUCACAGACAUUCGUGUCAGUUUUGCGCCAACCCAAACCGGUUUCGGCCAAAACCCC---------AUUACCGCCCGCUUCCUGGCCGCAGUUUCACCACAUGCAA .....((((...(((.....((((....)))).((((((.(((((((......))))..)))))))))...---------.........)))....)))).(((((......)).))).. ( -25.70) >DroYak_CAF1 31371 111 + 1 AAUUAGCCAAACGUGUCACAGACAUAUGUGUCAGUUUUGCGCCAACCCAAACCGGUUUCGGCCAAAACCCC---------AUUACCUCCCGCUUCCUGGCCGCAGUUUCACCACAUGCAA .....((((....((.((((......)))).))((((((.(((((((......))))..)))))))))...---------................)))).(((((......)).))).. ( -26.50) >consensus AAUUAGCCAAACGUGUCACAGACAUUUGUGUCAGUUUUGCGCCAACCCAAACCGGUUUCGGCCAAAACCCC_________AUUACCGCCCGCUUCCUGGCCGCAGUUUCACCACAUGCAA .....((((...(((.....((((....)))).((((((.(((((((......))))..))))))))).....................)))....)))).(((((......)).))).. (-25.76 = -25.57 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:21 2006