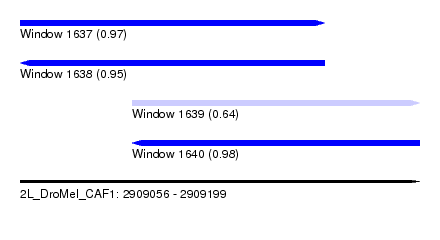
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,909,056 – 2,909,199 |
| Length | 143 |
| Max. P | 0.984176 |

| Location | 2,909,056 – 2,909,165 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 86.03 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -24.09 |
| Energy contribution | -23.87 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2909056 109 + 22407834 UUGUUGUUGGCAUCAGCUCAUUUAGCUGAUGCUACAUUUUCUUCUUGUUGUUGUUGCUGUUUUCCCUCUCUUUGGGGCUUUGUGGAAUAUAGACACCGAAUAAAACCAA ....(((.((((((((((.....)))))))))))))..........(((.(((.((((((.(((((.(((....)))....).)))).)))).)).)))....)))... ( -28.00) >DroEre_CAF1 28297 97 + 1 UUGUUUUUGGCAUCAGCUCAUUUAGCUGAUGCUACAUUUUG------------UUGUUGUUUUUCCCCUCUUUGGGGCUUUGAGGCAUAUAGACACCGAAUAAAACCGA ((((((.(((((((((((.....))))))))))).....((------------((..((((((.((((.....))))....))))))....))))..))))))...... ( -30.70) >DroYak_CAF1 28580 96 + 1 UUGUUGUUGGCAUCAGCUCAUUUAGCUGAUGCUACAUUUUG------------UUGUUGUU-UUCCCCUCUUUGGGGCUUUGUGGAAUAUAGACAGCGAAUAAAACCGA ....(((.((((((((((.....)))))))))))))(((((------------((((((((-(.((((.....))))((....)).....))))))).))))))).... ( -31.50) >consensus UUGUUGUUGGCAUCAGCUCAUUUAGCUGAUGCUACAUUUUG____________UUGUUGUUUUUCCCCUCUUUGGGGCUUUGUGGAAUAUAGACACCGAAUAAAACCGA .((((..(((((((((((.....)))))))))))..............................((((.....))))..............)))).............. (-24.09 = -23.87 + -0.22)

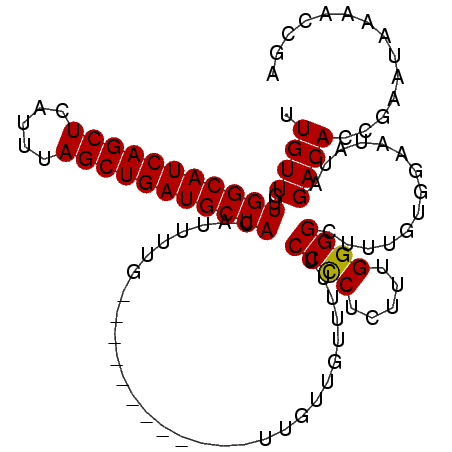
| Location | 2,909,056 – 2,909,165 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 86.03 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -19.69 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2909056 109 - 22407834 UUGGUUUUAUUCGGUGUCUAUAUUCCACAAAGCCCCAAAGAGAGGGAAAACAGCAACAACAACAAGAAGAAAAUGUAGCAUCAGCUAAAUGAGCUGAUGCCAACAACAA ((((((((.(((..(((...............(((........)))...............))).))).)))))...(((((((((.....)))))))))...)))... ( -23.01) >DroEre_CAF1 28297 97 - 1 UCGGUUUUAUUCGGUGUCUAUAUGCCUCAAAGCCCCAAAGAGGGGAAAAACAACAA------------CAAAAUGUAGCAUCAGCUAAAUGAGCUGAUGCCAAAAACAA ...(((((....(((((....)))))......((((.....)))).))))).....------------.........(((((((((.....)))))))))......... ( -27.10) >DroYak_CAF1 28580 96 - 1 UCGGUUUUAUUCGCUGUCUAUAUUCCACAAAGCCCCAAAGAGGGGAA-AACAACAA------------CAAAAUGUAGCAUCAGCUAAAUGAGCUGAUGCCAACAACAA ...(((((......(((.........)))...((((.....))))))-))).....------------.....(((.(((((((((.....)))))))))..))).... ( -23.80) >consensus UCGGUUUUAUUCGGUGUCUAUAUUCCACAAAGCCCCAAAGAGGGGAAAAACAACAA____________CAAAAUGUAGCAUCAGCUAAAUGAGCUGAUGCCAACAACAA .(((......))).(((...............((((.....))))................................(((((((((.....))))))))).....))). (-19.69 = -19.80 + 0.11)

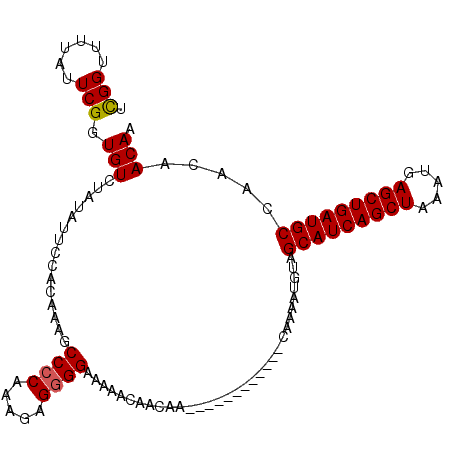
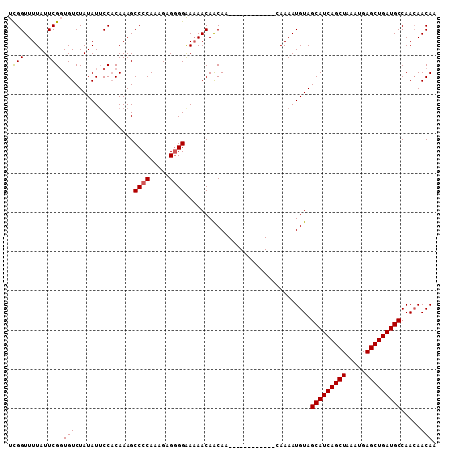
| Location | 2,909,096 – 2,909,199 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.84 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -18.23 |
| Energy contribution | -17.90 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2909096 103 + 22407834 CUUCUUGUUGUUGUUGCUGUUUUCCCUCUCUUUGGGGCUUUGUGGAAUAUAGACACCGAAUAAAACCAAAAAGAGGGAAAAUACCCAAAACUUAUCGAGUUUA ...((((..(((...(.((((((((((((.(((((.......(((..........))).......))))).)))))))))))).)...)))....)))).... ( -28.44) >DroEre_CAF1 28337 91 + 1 G------------UUGUUGUUUUUCCCCUCUUUGGGGCUUUGAGGCAUAUAGACACCGAAUAAAACCGAAAAGAGGGAAAACACCCAAAACUUAUCGAGUUUA (------------((..((((((.((((.....))))....))))))....)))........(((((((.(((.(((......)))....))).))).)))). ( -21.70) >DroYak_CAF1 28620 90 + 1 G------------UUGUUGUU-UUCCCCUCUUUGGGGCUUUGUGGAAUAUAGACAGCGAAUAAAACCGAAAAGAGGGAGAAUACCCGAAACUUAUCGAGUUUA .------------((((((((-(.((((.....))))((....)).....)))))))))...(((((((.(((.(((......)))....))).))).)))). ( -23.30) >consensus G____________UUGUUGUUUUUCCCCUCUUUGGGGCUUUGUGGAAUAUAGACACCGAAUAAAACCGAAAAGAGGGAAAAUACCCAAAACUUAUCGAGUUUA .................(((((((((.((.(((((...((((..(........)..)))).....))))).)).)))))))))....((((((...)))))). (-18.23 = -17.90 + -0.33)

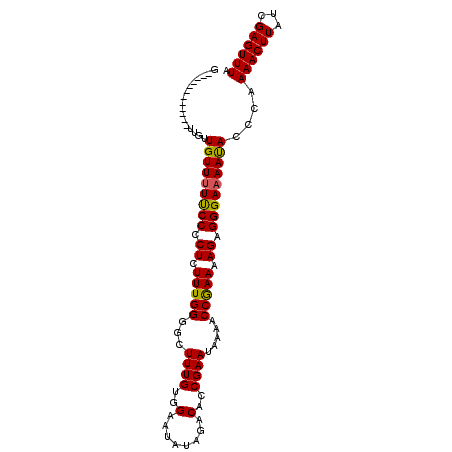
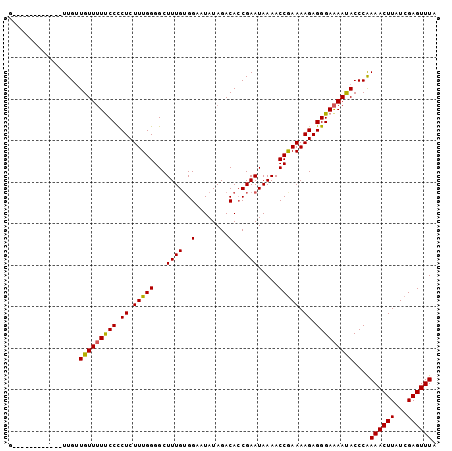
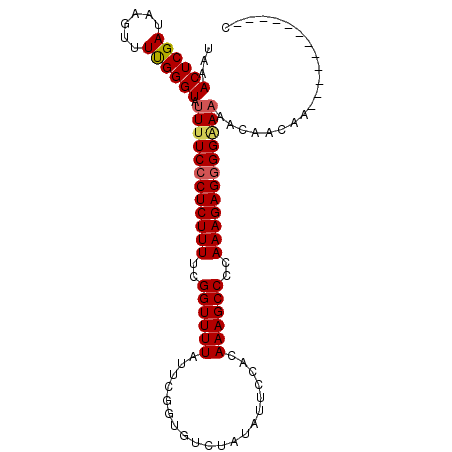
| Location | 2,909,096 – 2,909,199 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.84 |
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -20.25 |
| Energy contribution | -20.92 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2909096 103 - 22407834 UAAACUCGAUAAGUUUUGGGUAUUUUCCCUCUUUUUGGUUUUAUUCGGUGUCUAUAUUCCACAAAGCCCCAAAGAGAGGGAAAACAGCAACAACAACAAGAAG ...((((((......)))))).((((((((((((((((..((.....(((.........)))..))..))))))))))))))))................... ( -28.00) >DroEre_CAF1 28337 91 - 1 UAAACUCGAUAAGUUUUGGGUGUUUUCCCUCUUUUCGGUUUUAUUCGGUGUCUAUAUGCCUCAAAGCCCCAAAGAGGGGAAAAACAACAA------------C ...((((((......)))))).((((((((((((..((((((....(((((....)))))..))))))..))))))))))))........------------. ( -28.40) >DroYak_CAF1 28620 90 - 1 UAAACUCGAUAAGUUUCGGGUAUUCUCCCUCUUUUCGGUUUUAUUCGCUGUCUAUAUUCCACAAAGCCCCAAAGAGGGGAA-AACAACAA------------C ...((((((......)))))).(((.((((((((..((((((....................))))))..)))))))))))-........------------. ( -24.25) >consensus UAAACUCGAUAAGUUUUGGGUAUUUUCCCUCUUUUCGGUUUUAUUCGGUGUCUAUAUUCCACAAAGCCCCAAAGAGGGGAAAAACAACAA____________C ...((((((......)))))).((((((((((((..((((((....................))))))..))))))))))))..................... (-20.25 = -20.92 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:16 2006