| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,903,286 – 2,903,465 |
| Length | 179 |
| Max. P | 0.905688 |

| Location | 2,903,286 – 2,903,405 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.14 |
| Mean single sequence MFE | -20.46 |
| Consensus MFE | -14.10 |
| Energy contribution | -13.22 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2903286 119 + 22407834 UUUGGACAUCUCCAUCCUAUUACUAUGAUAUUGAAACAAUGUAUAUGAUGUGCUUUAUUUAUUUAAUAUUUGAAGCAUUCCAUUGCACAUUCCCAUCCCUAAUCUGUGCAUUCCAUUGC ..((((....))))......................(((((...(((..((((((((..(((...)))..))))))))..)))((((((...............))))))...))))). ( -21.46) >DroSec_CAF1 21907 119 + 1 UUCGGAAAUUACCAUCCUAUUACUAUGAUAUUGAAACAAUGCAUGUGAUGUGCUUUAUUUAUUUAAUAUCUGAAGCAUUCUAUUGCACAUUCCCAUCCCUAAUCUGUGCAUUCCACUGC ...((((...........(((((.(((.(((((...)))))))))))))((((((((..(((...)))..)))))))).....((((((...............))))))))))..... ( -22.16) >DroSim_CAF1 22318 119 + 1 UUCGGAAAUUACCAUCCUAUUACUAUGAUAUUGAAACAAUGUAUGUGAUGUGCUUUAUUUAUUUAAUAUUUGAAGCAUUCUAUUGCACAUUCCCAUCCCUAAUCUGUGCAUUCCACUGC ...((((...........(((((....((((((...))))))..)))))((((((((..(((...)))..)))))))).....((((((...............))))))))))..... ( -22.26) >DroEre_CAF1 22520 112 + 1 UUCGCA--UGAGCACUCUCCUAC-----AAUUGAAACAAUGUAUAUGAUGUGCUUUAUUUAUUUAAUAUUUGAAGCGUUCUAUUGCACAUUCCCAUCUCUAAUCUGUGCAUUCCACUGC ...(((--.((((((((...(((-----(.(((...)))))))...)).))((((((..(((...)))..))))))))))...((((((...............))))))......))) ( -18.66) >DroYak_CAF1 22658 112 + 1 UUCGCA--UGAGUUUUUUCCUAC-----UCUUAAAACAAUGUAUAUGAUGUGCUUUAUUUAUUUAAUUUUCGAAGCAUUCCAUUGCACAUUUCCAUCUCUAAUCUGUGCAUUCCACUGC ...(((--(((((........))-----)).......(((((..(((..(((((((...............)))))))..)))...)))))..............)))).......... ( -17.76) >consensus UUCGGA_AUGACCAUCCUAUUACUAUGAUAUUGAAACAAUGUAUAUGAUGUGCUUUAUUUAUUUAAUAUUUGAAGCAUUCUAUUGCACAUUCCCAUCCCUAAUCUGUGCAUUCCACUGC ...((......))...............................(((..((((((((..(((...)))..))))))))..)))((((((...............))))))......... (-14.10 = -13.22 + -0.88)

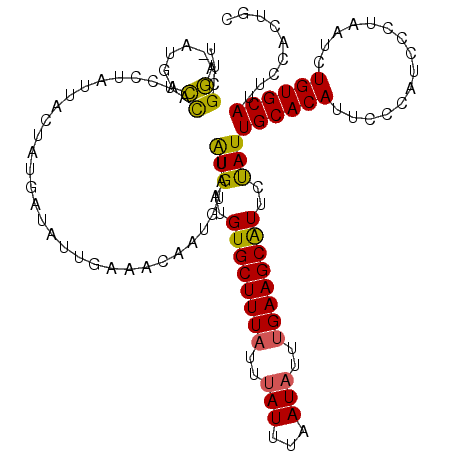
| Location | 2,903,366 – 2,903,465 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -15.34 |
| Energy contribution | -15.98 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2903366 99 + 22407834 CAUUGCACAUUCCCAUCCCUAAUCUGUGCAUUCCAUUGCCACUUUUUGUGU--GUC-GCUU-------------CCCGAAAGGAUUCUUU-UGGGGCAUGGAGCAUGGAGC---UAGCU ..................(((.(((((((..(((((.(((((.....))).--...-....-------------((((((((....))))-))))))))))))))))))..---))).. ( -28.40) >DroSec_CAF1 21987 94 + 1 UAUUGCACAUUCCCAUCCCUAAUCUGUGCAUUCCACUGCCACUUUUUGUGU--GUC-GCUC-------------CCCAAAAGGAUUCUUU-UUGG-----GGGCAUGGAUC---UAGCA ...((((((...............)))))).((((..(((((.....))).--)).-((.(-------------(((((((((...))))-))))-----)))).))))..---..... ( -28.46) >DroSim_CAF1 22398 95 + 1 UAUUGCACAUUCCCAUCCCUAAUCUGUGCAUUCCACUGCCACUUUUUGUGU--GUC-GCUC-------------CCCGAAAGGAUUCUUUUUUGG-----GGGCAUGGAGC---UAGCU ...((((((...............))))))(((((..(((((.....))).--)).-((.(-------------(((((((((....))))))))-----)))).))))).---..... ( -27.76) >DroEre_CAF1 22593 109 + 1 UAUUGCACAUUCCCAUCUCUAAUCUGUGCAUUCCACUGCCACUUUUUGUGU--GUGUGCCCCAUUGCCAGUCGGUUCGAAAGGAUUCUUU-U--G-----GGGCAUGGUGCAUGGUGCA ...(((((.................(((((((.....(((((.....))).--))((((((((.....((...((((....)))).))..-)--)-----))))))))))))).))))) ( -31.07) >DroYak_CAF1 22731 104 + 1 CAUUGCACAUUUCCAUCUCUAAUCUGUGCAUUCCACUGCCACUUUUUGUGUGUGUGUGCCCCAUUGCCAGUCGGUCCGAAAGGAUUCUUU-U--G-----GGGCAUGU-------UGCA ...((((((...............))))))...(((.(((((.....))).)))))(((((((.....((...((((....)))).))..-)--)-----)))))...-------.... ( -30.56) >consensus UAUUGCACAUUCCCAUCCCUAAUCUGUGCAUUCCACUGCCACUUUUUGUGU__GUC_GCUC_____________CCCGAAAGGAUUCUUU_U_GG_____GGGCAUGGAGC___UAGCA ...((((((...............)))))).((((.((((((.....))..........................((....))..................)))))))).......... (-15.34 = -15.98 + 0.64)

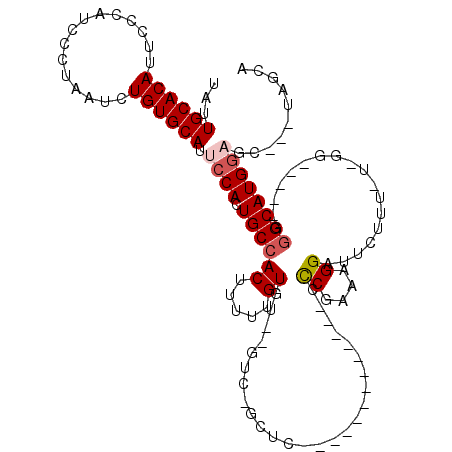
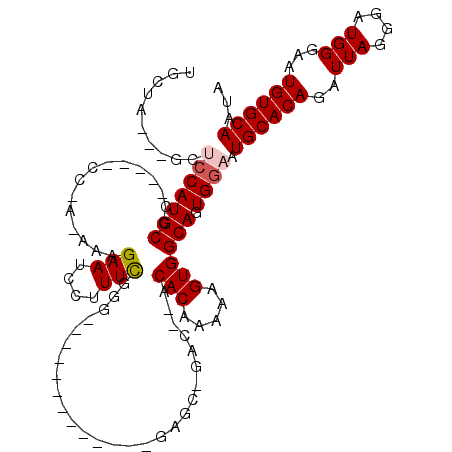
| Location | 2,903,366 – 2,903,465 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -14.88 |
| Energy contribution | -15.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2903366 99 - 22407834 AGCUA---GCUCCAUGCUCCAUGCCCCA-AAAGAAUCCUUUCGGG-------------AAGC-GAC--ACACAAAAAGUGGCAAUGGAAUGCACAGAUUAGGGAUGGGAAUGUGCAAUG .....---..(((((((....((((((.-((((....)))).)))-------------..))-)..--.(((.....))))).))))).((((((..(((....)))...))))))... ( -25.50) >DroSec_CAF1 21987 94 - 1 UGCUA---GAUCCAUGCCC-----CCAA-AAAGAAUCCUUUUGGG-------------GAGC-GAC--ACACAAAAAGUGGCAGUGGAAUGCACAGAUUAGGGAUGGGAAUGUGCAAUA .....---..(((((((((-----((((-((.......)))))))-------------).))-...--.(((.....)))...))))).((((((..(((....)))...))))))... ( -29.60) >DroSim_CAF1 22398 95 - 1 AGCUA---GCUCCAUGCCC-----CCAAAAAAGAAUCCUUUCGGG-------------GAGC-GAC--ACACAAAAAGUGGCAGUGGAAUGCACAGAUUAGGGAUGGGAAUGUGCAAUA .....---..(((((((((-----((...((((....)))).)))-------------).))-...--.(((.....)))...))))).((((((..(((....)))...))))))... ( -26.40) >DroEre_CAF1 22593 109 - 1 UGCACCAUGCACCAUGCCC-----C--A-AAAGAAUCCUUUCGAACCGACUGGCAAUGGGGCACAC--ACACAAAAAGUGGCAGUGGAAUGCACAGAUUAGAGAUGGGAAUGUGCAAUA ....((((((....(((((-----(--(-...(((....)))...((....))...)))))))...--.(((.....)))))).)))..((((((..(((....)))...))))))... ( -31.10) >DroYak_CAF1 22731 104 - 1 UGCA-------ACAUGCCC-----C--A-AAAGAAUCCUUUCGGACCGACUGGCAAUGGGGCACACACACACAAAAAGUGGCAGUGGAAUGCACAGAUUAGAGAUGGAAAUGUGCAAUG ....-------...(((((-----(--(-......(((....)))((....))...)))))))(((...(((.....)))...)))...((((((..((.......))..))))))... ( -27.70) >consensus UGCUA___GCUCCAUGCCC_____CC_A_AAAGAAUCCUUUCGGG_____________GAGC_GAC__ACACAAAAAGUGGCAGUGGAAUGCACAGAUUAGGGAUGGGAAUGUGCAAUA ..........(((((((...............(((....)))...........................(((.....)))))).)))).((((((..(((....)))...))))))... (-14.88 = -15.32 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:09 2006