| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,896,180 – 2,896,291 |
| Length | 111 |
| Max. P | 0.752879 |

| Location | 2,896,180 – 2,896,291 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.41 |
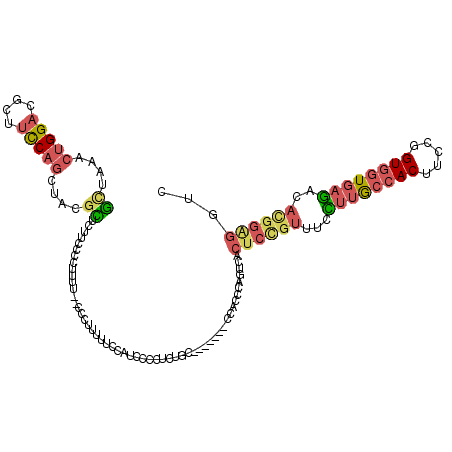
| Mean single sequence MFE | -26.44 |
| Consensus MFE | -15.78 |
| Energy contribution | -16.48 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
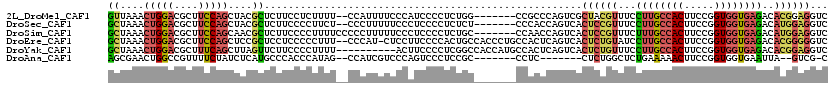
>2L_DroMel_CAF1 2896180 111 + 22407834 GUUAAACUGGACGCUUCCAGCUACGCUCUUCCUCUUUU--CCAUUUUCCCAUCCCCUCUGG-------CCGCCCAGUCGCUACGUUUCCUUGCCACUUCCGGUGGUGAGACACGGAGGUC ......(((((....)))))..................--........((.(((..(((.(-------(((((.(((.((...........)).)))...)))))).)))...))))).. ( -27.10) >DroSec_CAF1 14766 111 + 1 GCUAAACUGGACGCUUCCAGCUACGCUCUUCCCCUUCU--CCCUUUUUCCCUCCCCUCUCU-------CCCACCAGUCACUCCGUUUCCUUGCCACUUCCGGUGGUGAGACAUGGAGGUC ((....(((((....)))))....))............--.........(((((....(((-------((((((.(......((......))......).))))).))))...))))).. ( -27.40) >DroSim_CAF1 15164 113 + 1 GCUAAACUGGACGCUUCCAGCAACGCUCUUCCCCUUUUCCCCCUUUUUCCCUCCCCUCUGC-------CCAACCAGUCACUCCGUUUCUUUGCCACUUCCGGUGGUGAGACAUGGAGGUC ((....(((((....)))))....)).......................(((((...(((.-------.....))).......(((((...(((((.....))))))))))..))))).. ( -26.30) >DroEre_CAF1 15234 117 + 1 GCUAAACUGGACGCUUCCAGCUCCGCUCCUCCCCCUUU--CCCAU-CUCCUUCCCCACUGCCACCCUGCCACUCAGUCACUCUGUAUCCUUGCCACUUCCGGUGGUGAGACACGGGGGUC ((....(((((....)))))....))....(((((...--.....-...........................(((.....)))....((..((((.....))))..))....))))).. ( -29.60) >DroYak_CAF1 15331 110 + 1 GCUAAACUGGACGCUUUCAGCUUAGUUCUUCCCCUUUU----------ACUUCCCCUCGGCCACCAUGCCACUCAGUCACUCUGUUUCCUUGCCACUUCCGGUGGUGAGACACGGAGGUC (((((.(((((....))))).)))))............----------..........(((......))).........((((((...((..((((.....))))..))..))))))... ( -29.80) >DroAna_CAF1 11403 101 + 1 AGCGAACUGGCCGUUUUCUAUCUCAUGCCCACCCAUAG--CCAUCGUCCCAGUCCCUCCGC-------CCUC-------CUCUGGCUCUGAAAAACUUCCGGUGGUGAAUUA--GUCG-C .((((.((((.((....((((.............))))--....))..))))((((.((((-------(...-------....)))...(((....))).)).)).))....--.)))-) ( -18.42) >consensus GCUAAACUGGACGCUUCCAGCUACGCUCUUCCCCUUUU__CCCUUUUUCCAUCCCCUCUGC_______CCACCCAGUCACUCCGUUUCCUUGCCACUUCCGGUGGUGAGACACGGAGGUC ((....(((((....)))))....)).....................................................((((((...((((((((.....))))))))..))))))... (-15.78 = -16.48 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:57 2006