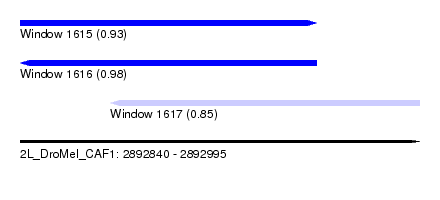
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,892,840 – 2,892,995 |
| Length | 155 |
| Max. P | 0.976678 |

| Location | 2,892,840 – 2,892,955 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.53 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -26.00 |
| Energy contribution | -26.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
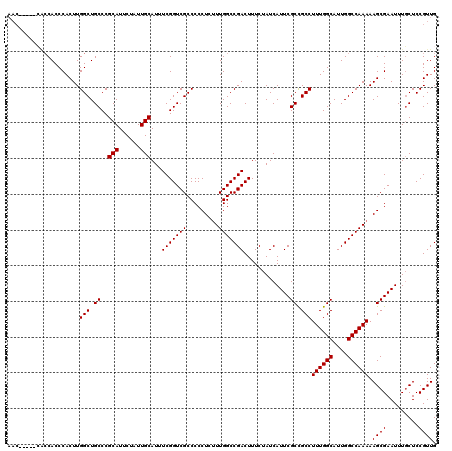
>2L_DroMel_CAF1 2892840 115 + 22407834 AAC-----CACCACCCACUUGGCUGCCCGCAUUCUAUUGCAUUUCGGUCGCCCCCUCUUUGGCCGACUUUCUAUCAUUCGCCGCCUUUGGCAUUGGCCAAAAAGCGAAUUUGCUCCGUUG ...-----............(((.((..(((......)))...(((((((.........))))))).............)).)))((((((....)))))).((((....))))...... ( -26.40) >DroSec_CAF1 11521 115 + 1 AAC-----CACCACCCACUUGGCUGCCCGCAUUCUAUUGCAUUUCGGUCGCCCCCUCUUUGGCCGACUUUCUAUCAUUCGCCGCCUUUGGCAUUGGCCAAAAAGCGAAUUUGCUCCGUUG ...-----............(((.((..(((......)))...(((((((.........))))))).............)).)))((((((....)))))).((((....))))...... ( -26.40) >DroSim_CAF1 11843 115 + 1 AAC-----CGCCACCCACUUGGCUGCCCGCAUUCUAUUGCAUUUCGGUCGCCCCCUCUUUGGCCGACUUUCUAUCAUUCGCCGCCUUUGGCAUUGGCCAAAAAGCGAAUUUGCUCCGUUG ...-----.((((......)))).((..(((......)))...(((((((.........))))))).........((((((....((((((....))))))..))))))..))....... ( -29.00) >DroEre_CAF1 11737 120 + 1 AACAUGCCAACCACCCACUUGGCUGCCCGCAUUCUAUUGCAUUUCGGUCGCCCCCUCGUUGGCCGACUUUCUAUCAUUCGCUGCCUUUGGCAUUGGCCAAAAAGCGAAUUUUCUCCGUUG .....((((((.........(((.(((.(((......))).....))).))).....))))))((((........(((((((...((((((....)))))).))))))).......)))) ( -33.20) >DroYak_CAF1 11848 109 + 1 -----------CACCCACUUGGCUGCCCGCAUUCUAUUGCAUUUCGGUCGCCCCCUCUUUGGCCGACUUUCUAUCAUUCGCUGCCUUUGGCAUUGGCCAAAAAGCGAAUUUGCUCCGUUG -----------.........((..((..(((......)))...(((((((.........))))))).........(((((((...((((((....)))))).)))))))..)).)).... ( -29.00) >consensus AAC_____CACCACCCACUUGGCUGCCCGCAUUCUAUUGCAUUUCGGUCGCCCCCUCUUUGGCCGACUUUCUAUCAUUCGCCGCCUUUGGCAUUGGCCAAAAAGCGAAUUUGCUCCGUUG ....................(((.((..(((......)))...(((((((.........))))))).............)).)))((((((....)))))).((((.........)))). (-26.00 = -26.00 + 0.00)

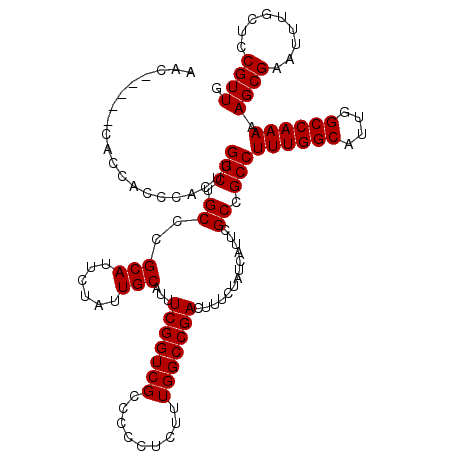
| Location | 2,892,840 – 2,892,955 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.53 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -32.58 |
| Energy contribution | -32.74 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2892840 115 - 22407834 CAACGGAGCAAAUUCGCUUUUUGGCCAAUGCCAAAGGCGGCGAAUGAUAGAAAGUCGGCCAAAGAGGGGGCGACCGAAAUGCAAUAGAAUGCGGGCAGCCAAGUGGGUGGUG-----GUU .......(((..(((.((((((((((..((((......))))...(((.....))))))))))))).((....))))).)))........((.(.((.((....)).)).).-----)). ( -36.40) >DroSec_CAF1 11521 115 - 1 CAACGGAGCAAAUUCGCUUUUUGGCCAAUGCCAAAGGCGGCGAAUGAUAGAAAGUCGGCCAAAGAGGGGGCGACCGAAAUGCAAUAGAAUGCGGGCAGCCAAGUGGGUGGUG-----GUU .......(((..(((.((((((((((..((((......))))...(((.....))))))))))))).((....))))).)))........((.(.((.((....)).)).).-----)). ( -36.40) >DroSim_CAF1 11843 115 - 1 CAACGGAGCAAAUUCGCUUUUUGGCCAAUGCCAAAGGCGGCGAAUGAUAGAAAGUCGGCCAAAGAGGGGGCGACCGAAAUGCAAUAGAAUGCGGGCAGCCAAGUGGGUGGCG-----GUU ...(((.((....((.((((((((((..((((......))))...(((.....))))))))))))).))))..)))...((((......))))(.((.((....)).)).).-----... ( -37.90) >DroEre_CAF1 11737 120 - 1 CAACGGAGAAAAUUCGCUUUUUGGCCAAUGCCAAAGGCAGCGAAUGAUAGAAAGUCGGCCAACGAGGGGGCGACCGAAAUGCAAUAGAAUGCGGGCAGCCAAGUGGGUGGUUGGCAUGUU .((((......((((((((((((((....)))))))..)))))))........((((((((.((....(((..((....((((......))))))..)))...))..)))))))).)))) ( -38.50) >DroYak_CAF1 11848 109 - 1 CAACGGAGCAAAUUCGCUUUUUGGCCAAUGCCAAAGGCAGCGAAUGAUAGAAAGUCGGCCAAAGAGGGGGCGACCGAAAUGCAAUAGAAUGCGGGCAGCCAAGUGGGUG----------- ...(((.((....((.((((((((((..((((...))))......(((.....))))))))))))).))))..)))...((((......))))..((.((....)).))----------- ( -33.20) >consensus CAACGGAGCAAAUUCGCUUUUUGGCCAAUGCCAAAGGCGGCGAAUGAUAGAAAGUCGGCCAAAGAGGGGGCGACCGAAAUGCAAUAGAAUGCGGGCAGCCAAGUGGGUGGUG_____GUU ...(((.((....((.((((((((((..((((...))))......(((.....))))))))))))).))))..)))...((((......))))..((.((....)).))........... (-32.58 = -32.74 + 0.16)

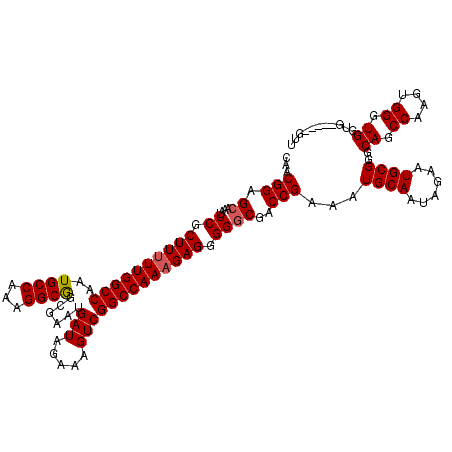
| Location | 2,892,875 – 2,892,995 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -29.94 |
| Energy contribution | -29.70 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
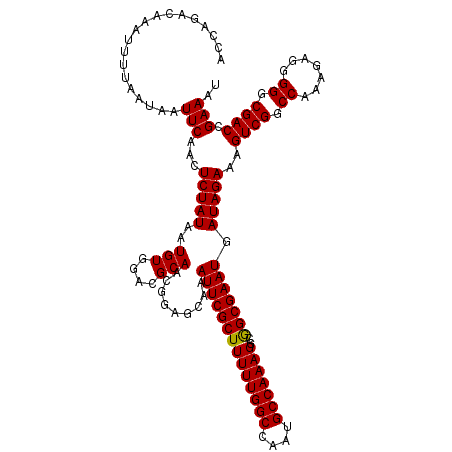
>2L_DroMel_CAF1 2892875 120 - 22407834 ACCAGACAAAUUUUAAUAAUUCAACUCUAUAAUGUGGACGCAACGGAGCAAAUUCGCUUUUUGGCCAAUGCCAAAGGCGGCGAAUGAUAGAAAGUCGGCCAAAGAGGGGGCGACCGAAAU ...................(((.((........))((.(((..((((.....))).((((((((((..((((......))))...(((.....))))))))))))))..))).))))).. ( -31.40) >DroSec_CAF1 11556 120 - 1 ACCAGACAAAUUUUAAUAAUUCAACUCUAUAAUGUGGACGCAACGGAGCAAAUUCGCUUUUUGGCCAAUGCCAAAGGCGGCGAAUGAUAGAAAGUCGGCCAAAGAGGGGGCGACCGAAAU ...................(((.((........))((.(((..((((.....))).((((((((((..((((......))))...(((.....))))))))))))))..))).))))).. ( -31.40) >DroSim_CAF1 11878 120 - 1 ACCAGACAAAUUUUAAUAAUUCAACUCUAUAAUGUGGACGCAACGGAGCAAAUUCGCUUUUUGGCCAAUGCCAAAGGCGGCGAAUGAUAGAAAGUCGGCCAAAGAGGGGGCGACCGAAAU ...................(((.((........))((.(((..((((.....))).((((((((((..((((......))))...(((.....))))))))))))))..))).))))).. ( -31.40) >DroEre_CAF1 11777 120 - 1 ACCAGACAAAUUUUAAUAAUUCAACUCUAUAAUGUGGACGCAACGGAGAAAAUUCGCUUUUUGGCCAAUGCCAAAGGCAGCGAAUGAUAGAAAGUCGGCCAACGAGGGGGCGACCGAAAU ...................(((...(((((..(((....))).........((((((((((((((....)))))))..))))))).)))))..((((.((.......)).)))).))).. ( -29.70) >DroYak_CAF1 11877 120 - 1 ACCAGACAAAUUUUAAUAAUUCAACUCUAUAAUGUGGACGCAACGGAGCAAAUUCGCUUUUUGGCCAAUGCCAAAGGCAGCGAAUGAUAGAAAGUCGGCCAAAGAGGGGGCGACCGAAAU ...................(((..((((.....((.(((((......))..((((((((((((((....)))))))..)))))))........))).))...)))).((....))))).. ( -31.30) >consensus ACCAGACAAAUUUUAAUAAUUCAACUCUAUAAUGUGGACGCAACGGAGCAAAUUCGCUUUUUGGCCAAUGCCAAAGGCGGCGAAUGAUAGAAAGUCGGCCAAAGAGGGGGCGACCGAAAU ...................(((...(((((..(((....))).........((((((((((((((....)))))))..))))))).)))))..((((.((.......)).)))).))).. (-29.94 = -29.70 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:54 2006