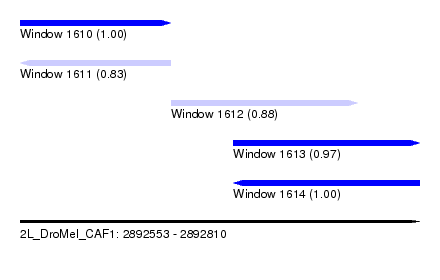
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,892,553 – 2,892,810 |
| Length | 257 |
| Max. P | 0.999393 |

| Location | 2,892,553 – 2,892,650 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 91.18 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -21.06 |
| Energy contribution | -21.26 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.999393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
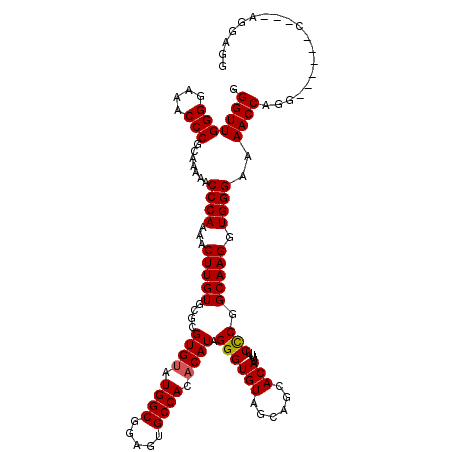
>2L_DroMel_CAF1 2892553 97 + 22407834 ACGUGUGCGACACUCUCAACAAACUGAUAAAAUUUUAAUUUCUCCAAAAUUGAAAUUUUAAUGUGUGUCUAGAUCGCGGUUUACCCCCGUUCAG-------CAC ....(((((((((.((((......)))((((((((((((((.....))))))))))))))..).)))))..((.((.((......)))).)).)-------))) ( -24.40) >DroSec_CAF1 11243 97 + 1 ACGUGUGCGACACUCUCAACAAACUGAUAAAAUUUUAAUUUCUCCAAAAUUGAAAUUUUAAUGUGUGUCUAGAGCGCGGUUUACCCCCGUUCAG-------CAC ....(((((((((.((((......)))((((((((((((((.....))))))))))))))..).)))))..(((((.((......))))))).)-------))) ( -28.50) >DroSim_CAF1 11551 97 + 1 ACGUGUGCGACACUCUCAACAAACUGAUAAAAUUUUAAUUUCUCCAAAAUUGAAAUUUUAAUGUGUGUCUAGAGCGCGGUUUACCCCCGUUCAG-------CAC ....(((((((((.((((......)))((((((((((((((.....))))))))))))))..).)))))..(((((.((......))))))).)-------))) ( -28.50) >DroEre_CAF1 11490 87 + 1 ACGUGUGCGACACUCUCAACAAACUGAUAAAAUUUUAAUUUCUCCAAAAUUGAAAUUUUAAUGUGUGUCUAGAGCGCGGUUUACCU-----------------C .(((((..(((((.((((......)))((((((((((((((.....))))))))))))))..).)))))....)))))........-----------------. ( -21.80) >DroYak_CAF1 11589 104 + 1 ACGUGUGCGACACUCUCAACAAACUGAUAAAAUUUUAAUUUCUCCAAAAUUGAAAUUUUAAUGUGUGUCUAGAGCGCGGUUUACCCCUGUUCCAAGGCUCCCCC .(((((..(((((.((((......)))((((((((((((((.....))))))))))))))..).)))))....))))).......(((......)))....... ( -23.90) >consensus ACGUGUGCGACACUCUCAACAAACUGAUAAAAUUUUAAUUUCUCCAAAAUUGAAAUUUUAAUGUGUGUCUAGAGCGCGGUUUACCCCCGUUCAG_______CAC .(((((..(((((.((((......)))((((((((((((((.....))))))))))))))..).)))))....))))).......................... (-21.06 = -21.26 + 0.20)

| Location | 2,892,553 – 2,892,650 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 91.18 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.52 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2892553 97 - 22407834 GUG-------CUGAACGGGGGUAAACCGCGAUCUAGACACACAUUAAAAUUUCAAUUUUGGAGAAAUUAAAAUUUUAUCAGUUUGUUGAGAGUGUCGCACACGU (((-------(.((.((.((.....)).)).))..(((((....((((((((.((((((...)))))).))))))))((((....))))..))))))))).... ( -26.00) >DroSec_CAF1 11243 97 - 1 GUG-------CUGAACGGGGGUAAACCGCGCUCUAGACACACAUUAAAAUUUCAAUUUUGGAGAAAUUAAAAUUUUAUCAGUUUGUUGAGAGUGUCGCACACGU (((-------(......((......))(((((((.((((.((..((((((((.((((((...)))))).))))))))...)).)))).))))))).)))).... ( -27.40) >DroSim_CAF1 11551 97 - 1 GUG-------CUGAACGGGGGUAAACCGCGCUCUAGACACACAUUAAAAUUUCAAUUUUGGAGAAAUUAAAAUUUUAUCAGUUUGUUGAGAGUGUCGCACACGU (((-------(......((......))(((((((.((((.((..((((((((.((((((...)))))).))))))))...)).)))).))))))).)))).... ( -27.40) >DroEre_CAF1 11490 87 - 1 G-----------------AGGUAAACCGCGCUCUAGACACACAUUAAAAUUUCAAUUUUGGAGAAAUUAAAAUUUUAUCAGUUUGUUGAGAGUGUCGCACACGU .-----------------.((....))(((((((.((((.((..((((((((.((((((...)))))).))))))))...)).)))).)))))))......... ( -20.60) >DroYak_CAF1 11589 104 - 1 GGGGGAGCCUUGGAACAGGGGUAAACCGCGCUCUAGACACACAUUAAAAUUUCAAUUUUGGAGAAAUUAAAAUUUUAUCAGUUUGUUGAGAGUGUCGCACACGU ((...(.(((((...))))).)...))(((((((.((((.((..((((((((.((((((...)))))).))))))))...)).)))).)))))))......... ( -27.00) >consensus GUG_______CUGAACGGGGGUAAACCGCGCUCUAGACACACAUUAAAAUUUCAAUUUUGGAGAAAUUAAAAUUUUAUCAGUUUGUUGAGAGUGUCGCACACGU ...................((....))(((((((.((((.((..((((((((.((((((...)))))).))))))))...)).)))).)))))))......... (-19.32 = -19.52 + 0.20)


| Location | 2,892,650 – 2,892,770 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.61 |
| Mean single sequence MFE | -26.04 |
| Consensus MFE | -19.40 |
| Energy contribution | -19.28 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2892650 120 + 22407834 CCCUUUUAUCCACCAACUCGACUGCACUUUGUGUUCUCCUCCUCCUCCUCCUGCUGCCUGGUUUUCCCACCUUGCCGGGAAAAUGUGCUGCUACACCCUAUGUGUGGCACUUCGCCAUAC ...................((..((((...))))..))..............((.((...((((((((........))))))))..)).))..........(((((((.....))))))) ( -25.90) >DroSec_CAF1 11340 111 + 1 CCCUUUUACCCACCAACUCGACUGCACUUUGUGUCCGCCUCCUCCU---G------CCUGGUUUUCCCACCUUGCCGGGAAAAUGUGCUGCUACACCCUAUGUGUGGCACUCCGCCAUAC .......................((((.........((........---)------)...((((((((........))))))))))))((((((((.....))))))))........... ( -25.20) >DroSim_CAF1 11648 115 + 1 CCCUUUUACCCACCAACUCGACUGCACUUUGUGUCCGCCUCCUCCU---CCUGC--CCUGGUUUUCCCACCUUGCCGGGAAAAUGUGCUGCUACACCCUAUGUGUGGCACUCCGCCAUAC .......................((((...((....))........---.....--....((((((((........))))))))))))((((((((.....))))))))........... ( -24.40) >DroEre_CAF1 11577 103 + 1 CCUACUUUCCCUCCAUUUCGACUGCACUUUGUGUCCACC-----CU---G------CCAGGUUUUCCCACCUUGCCGAGAAAAUGUG---CUACACCCUAUGUGUGGCACUCCGCCAUGC ...............(((((...((((...)))).....-----..---(------(.((((......)))).)))))))....(((---((((((.....))))))))).......... ( -24.10) >DroYak_CAF1 11693 103 + 1 CCCUUUUUCCCUGCAUUUCGACUGCACUUUGUGUCCUCC-----CU---G------CCCGGUUUUCCCACCUUGCCGGGAAAAUGUG---CUACACCCAAUGUGUGGCACUCCGCCAUAC ....(((((((.(((....((..((((...))))..)).-----..---.------...(((......))).))).))))))).(((---((((((.....))))))))).......... ( -30.60) >consensus CCCUUUUACCCACCAACUCGACUGCACUUUGUGUCCGCCUCCUCCU___G______CCUGGUUUUCCCACCUUGCCGGGAAAAUGUGCUGCUACACCCUAUGUGUGGCACUCCGCCAUAC ...................(((.((.....))))).........................((((((((........))))))))(((......))).....(((((((.....))))))) (-19.40 = -19.28 + -0.12)

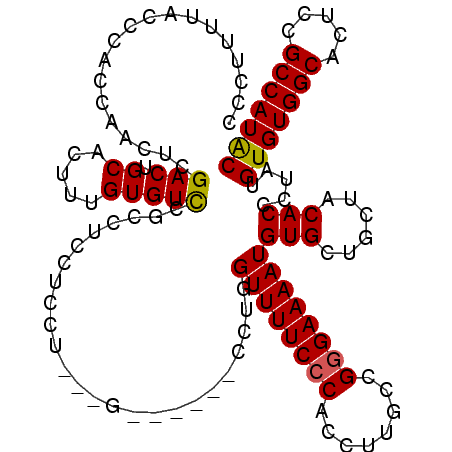
| Location | 2,892,690 – 2,892,810 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.66 |
| Mean single sequence MFE | -35.46 |
| Consensus MFE | -28.84 |
| Energy contribution | -28.88 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2892690 120 + 22407834 CCUCCUCCUCCUGCUGCCUGGUUUUCCCACCUUGCCGGGAAAAUGUGCUGCUACACCCUAUGUGUGGCACUUCGCCAUACACGCGCACAAGUUUUGGGUUUUUGCGGGUUUCCCCAACCC .........(((((.(((..((((((((........)))))))(((((((((((((.....))))))))....((.......)))))))....)..)))....)))))............ ( -39.30) >DroSec_CAF1 11380 111 + 1 CCUCCU---G------CCUGGUUUUCCCACCUUGCCGGGAAAAUGUGCUGCUACACCCUAUGUGUGGCACUCCGCCAUACACGCGCACAAGUUUUGGGUUUUUGCGGGUUUCCCCAACCC ......---(------((..((((((((........)))))))(((((((((((((.....))))))))....((.......)))))))....)..)))......(((((.....))))) ( -35.30) >DroSim_CAF1 11688 115 + 1 CCUCCU---CCUGC--CCUGGUUUUCCCACCUUGCCGGGAAAAUGUGCUGCUACACCCUAUGUGUGGCACUCCGCCAUACACGCGCACAAGUUUUGGGUUUUUGCGGGUUUCCCCAACCC ......---...((--((..((((((((........))))))))((((((((((((.....))))))))....((.......)))))).......))))......(((((.....))))) ( -36.70) >DroEre_CAF1 11616 104 + 1 ----CU---G------CCAGGUUUUCCCACCUUGCCGAGAAAAUGUG---CUACACCCUAUGUGUGGCACUCCGCCAUGCACGCGCACAAGUUUUGGGUUUUUGCGGGUUUCCCCAACCC ----..---(------(.((((......)))).(((.((((..((((---(.........((((((((.....))))))))...)))))..)))).)))....))(((((.....))))) ( -29.70) >DroYak_CAF1 11732 104 + 1 ----CU---G------CCCGGUUUUCCCACCUUGCCGGGAAAAUGUG---CUACACCCAAUGUGUGGCACUCCGCCAUACACGCGCACAAGUUCUGGGUUUUUGCGGGUUUCCCCAACCC ----..---(------((((((((((((........)))))))((((---(.........((((((((.....))))))))...)))))....))))))......(((((.....))))) ( -36.30) >consensus CCUCCU___G______CCUGGUUUUCCCACCUUGCCGGGAAAAUGUGCUGCUACACCCUAUGUGUGGCACUCCGCCAUACACGCGCACAAGUUUUGGGUUUUUGCGGGUUUCCCCAACCC ....................((((((((........))))))))..........((((..((((((((.....)))))))).((......))...))))......(((((.....))))) (-28.84 = -28.88 + 0.04)

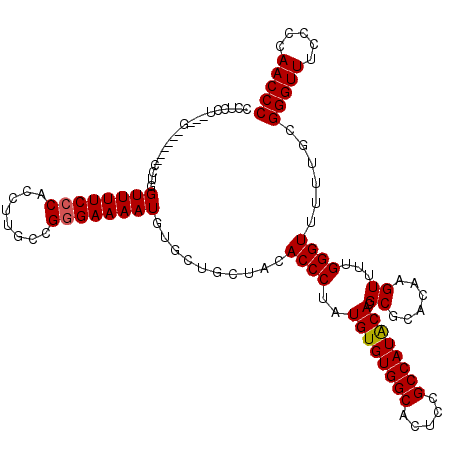
| Location | 2,892,690 – 2,892,810 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.66 |
| Mean single sequence MFE | -43.14 |
| Consensus MFE | -35.64 |
| Energy contribution | -35.68 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2892690 120 - 22407834 GGGUUGGGGAAACCCGCAAAAACCCAAAACUUGUGCGCGUGUAUGGCGAAGUGCCACACAUAGGGUGUAGCAGCACAUUUUCCCGGCAAGGUGGGAAAACCAGGCAGCAGGAGGAGGAGG ((((((((....))).....)))))....(((.(((((((((.((((.....)))).)))).((((((....)))).((((((((......))))))))))..)).))).)))....... ( -46.40) >DroSec_CAF1 11380 111 - 1 GGGUUGGGGAAACCCGCAAAAACCCAAAACUUGUGCGCGUGUAUGGCGGAGUGCCACACAUAGGGUGUAGCAGCACAUUUUCCCGGCAAGGUGGGAAAACCAGG------C---AGGAGG ((((((((....))).....)))))....(((.(((..((((.((((.....)))).)))).((((((....)))).((((((((......))))))))))..)------)---).))). ( -44.40) >DroSim_CAF1 11688 115 - 1 GGGUUGGGGAAACCCGCAAAAACCCAAAACUUGUGCGCGUGUAUGGCGGAGUGCCACACAUAGGGUGUAGCAGCACAUUUUCCCGGCAAGGUGGGAAAACCAGG--GCAGG---AGGAGG ((((((((....))).....)))))....(((.(((.(((((.((((.....)))).)))).((((((....)))).((((((((......)))))))))).).--))).)---)).... ( -46.30) >DroEre_CAF1 11616 104 - 1 GGGUUGGGGAAACCCGCAAAAACCCAAAACUUGUGCGCGUGCAUGGCGGAGUGCCACACAUAGGGUGUAG---CACAUUUUCUCGGCAAGGUGGGAAAACCUGG------C---AG---- ((((((((....))).......((((...(((((.(((.......)))..((((.((((.....)))).)---))).........))))).))))..)))))..------.---..---- ( -39.20) >DroYak_CAF1 11732 104 - 1 GGGUUGGGGAAACCCGCAAAAACCCAGAACUUGUGCGCGUGUAUGGCGGAGUGCCACACAUUGGGUGUAG---CACAUUUUCCCGGCAAGGUGGGAAAACCGGG------C---AG---- ((((((((....))).....)))))........(((.((.((........((((.((((.....)))).)---)))..(((((((......))))))))))).)------)---).---- ( -39.40) >consensus GGGUUGGGGAAACCCGCAAAAACCCAAAACUUGUGCGCGUGUAUGGCGGAGUGCCACACAUAGGGUGUAGCAGCACAUUUUCCCGGCAAGGUGGGAAAACCAGG______C___AGGAGG .(((((((....))).......((((...(((((....((((.((((.....)))).)))).((((((......)))....))).))))).))))..))))................... (-35.64 = -35.68 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:51 2006