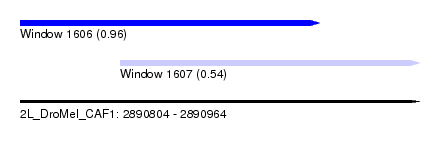
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,890,804 – 2,890,964 |
| Length | 160 |
| Max. P | 0.962216 |

| Location | 2,890,804 – 2,890,924 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.32 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -26.92 |
| Energy contribution | -26.68 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
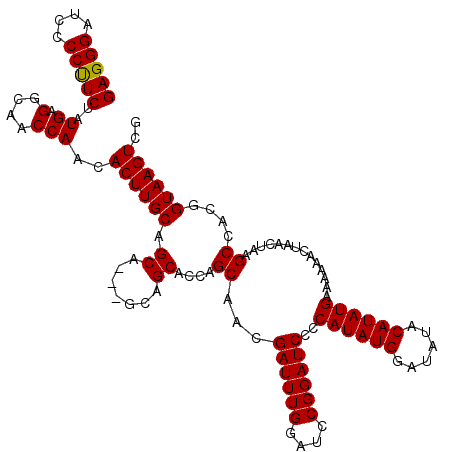
>2L_DroMel_CAF1 2890804 120 + 22407834 GAGGGAUCUCCUUCUAUGAGGCAACCAACACUUGCAGCAGCAGCAGCACCAGCAACGAUUUGGAUCCGGAUCCCCAUAUGGAUAUACAUAUGAAAAAACUAACUAAGCCACGGUAAGUCG (((((....))))).....(((.(((......(((.((....)).)))...((...((((((....))))))..((((((......))))))..............))...)))..))). ( -28.50) >DroSec_CAF1 9577 117 + 1 GAGGGAUCCCCUUCUAUGAGGCAACCAACACUUGCAGCA---GCAGCACCAGCAACGAUUUGGAUCCGGAUCCCCAUAUGGAUAUACAUAUGAAAAAACUAACUAAGCCACGGUAAGUUU ..(((((((((((....))))...((((....(((....---)))((....))......))))....)))))))((((((......))))))...(((((.(((.......))).))))) ( -30.70) >DroSim_CAF1 9795 117 + 1 GAGGGAUCCCCUUCUAUGAGGCAACCAACACUUGCAGCA---GCAGCACCAGCAACGAUUUGGAUUCGGAUCCCCAUAUGGAUAUACAUAUGAAAAAACUAACUAAGCCACGGUAAGUUU ..(((((((((((....))))...((((....(((....---)))((....))......))))....)))))))((((((......))))))...(((((.(((.......))).))))) ( -30.70) >DroEre_CAF1 9660 114 + 1 GAGGGAUCCCCCUCUAUGAGGCAACCAACACUUGCAGC------AGCACCAGCAACGAUUUGGAACCGGAUCCCCAUAUGGAUAUACAUAUGAAAAAACUAACUAAGCCACGGUAAGUCG (((((....)))))..((.(....)))..((((((...------.((.........((((((....))))))..((((((......))))))..............))....)))))).. ( -29.00) >DroYak_CAF1 9753 114 + 1 GAGGGAACACCCUCUAUGAGGCAACCAACACUUGCAGC------AGCACCAGCAACGAUUUGGAUCCGGAUCCCCAUAUGGAUAUACAUAUGAAAAAACUAACUAAGCCACGGUAAGUCG (((((....)))))..((.(....)))..((((((...------.((.........((((((....))))))..((((((......))))))..............))....)))))).. ( -28.60) >consensus GAGGGAUCCCCUUCUAUGAGGCAACCAACACUUGCAGCA___GCAGCACCAGCAACGAUUUGGAUCCGGAUCCCCAUAUGGAUAUACAUAUGAAAAAACUAACUAAGCCACGGUAAGUCG (((((....)))))..((.(....)))..((((((.((.......))....((...((((((....))))))..((((((......))))))..............))....)))))).. (-26.92 = -26.68 + -0.24)

| Location | 2,890,844 – 2,890,964 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -18.47 |
| Consensus MFE | -15.20 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
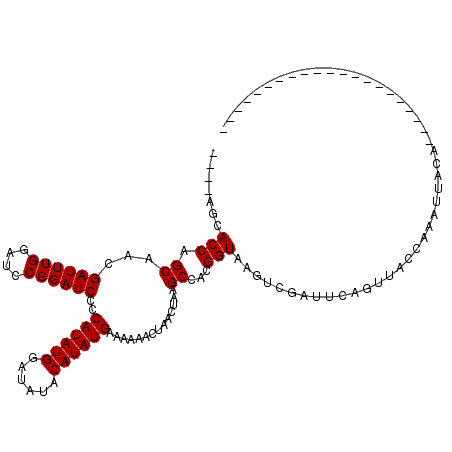
>2L_DroMel_CAF1 2890844 120 + 22407834 CAGCAGCACCAGCAACGAUUUGGAUCCGGAUCCCCAUAUGGAUAUACAUAUGAAAAAACUAACUAAGCCACGGUAAGUCGAUUCAGUUAACAAAUUUCACCUAAAGGAAUAUCCCACAUU .(((.((....))..((((((((((....)))).((((((......))))))......................)))))).....))).................((......))..... ( -17.80) >DroEre_CAF1 9698 95 + 1 ----AGCACCAGCAACGAUUUGGAACCGGAUCCCCAUAUGGAUAUACAUAUGAAAAAACUAACUAAGCCACGGUAAGUCGAUUCAGCUACCUACUUACC--------------------- ----.......((...((((((....))))))..((((((......))))))..............))...(((((((.(..........).)))))))--------------------- ( -19.10) >DroYak_CAF1 9791 95 + 1 ----AGCACCAGCAACGAUUUGGAUCCGGAUCCCCAUAUGGAUAUACAUAUGAAAAAACUAACUAAGCCACGGUAAGUCGAUUCAAUUACCAAAUUACU--------------------- ----.((....))...(((((((....(((((..((((((......)))))).....(((.(((.......))).))).))))).....)))))))...--------------------- ( -18.50) >consensus ____AGCACCAGCAACGAUUUGGAUCCGGAUCCCCAUAUGGAUAUACAUAUGAAAAAACUAACUAAGCCACGGUAAGUCGAUUCAGUUACCAAAUUACA_____________________ .......(((.((...((((((....))))))..((((((......))))))..............))...))).............................................. (-15.20 = -15.20 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:45 2006