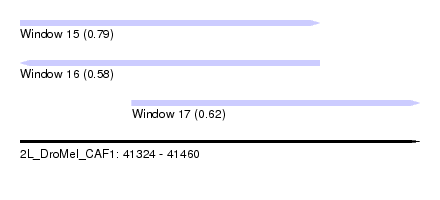
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 41,324 – 41,460 |
| Length | 136 |
| Max. P | 0.791148 |

| Location | 41,324 – 41,426 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.80 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -17.96 |
| Energy contribution | -17.68 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
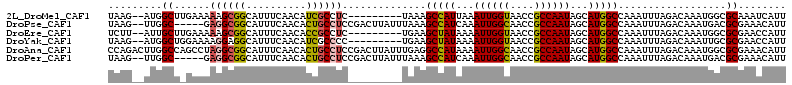
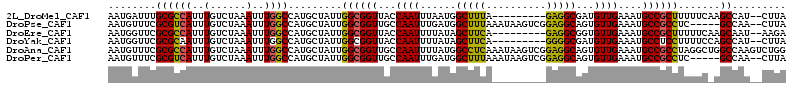
>2L_DroMel_CAF1 41324 102 + 22407834 UAAG--AUGGCUUGAAAAAGCGGCAUUUCAACAUCGCCUC---------UAAAGCCAUUAAAUUGGUAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGGCGCAAAUCAUU ...(--(((((((....))))((....))..))))(((((---------(((((((((...((((((....))))))...)))))....)))))).....))).......... ( -27.00) >DroPse_CAF1 21981 106 + 1 UAAG--UUGGC-----GAGGCGGCAUUUCAACACUGCCUCCGACUUAUUUAAAGCCAUCAAAUUGGCAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGACGCGAAACAUU ((((--((((.-----.((((((..........))))))))))))))(((((((((((...((((((....))))))...)))))....)))))).................. ( -30.60) >DroEre_CAF1 18920 102 + 1 UCUU--AUUGCUUGAAAAAGCGGCAUUUCAACACCGCCUC---------UGAAGCUAUAAAAUUGGUAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGGCGCGAACCAUU ....--.((((((....))))))..........(((((((---------(((((((((...((((((....))))))...)))))....)))))).....)))).)....... ( -24.00) >DroYak_CAF1 18183 102 + 1 UAAG--AUGGCUGGAAAAGGAGGCAUUUCAACAUCGCCCC---------UGAAGCUAUAAAAUUGGUAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUUGCGCGAACCAUU ....--.(((((.....(((.(((...........)))))---------)...(((((.....((((....)))))))))..))))).......................... ( -21.70) >DroAna_CAF1 20662 113 + 1 CCAGACUUGGCCAGCCUAGGCGGCAUUUCAACACUGCCUCCGACUUAUUUGAGGCCAUAAAAUUGGCAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGGCGCGAAACAUU ......(((((((..(((((.((((.........)))).))...........((((((...((((((....))))))...)))))).....))).....)))).)))...... ( -33.70) >DroPer_CAF1 21741 106 + 1 UAAG--UUGGC-----GAGGCGGCAUUUCAACACUGCCUCCGACUUAUUUAAAGCCAUCAAAUUGGCAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGACGCGAAACAUU ((((--((((.-----.((((((..........))))))))))))))(((((((((((...((((((....))))))...)))))....)))))).................. ( -30.60) >consensus UAAG__AUGGCU_GAAAAGGCGGCAUUUCAACACCGCCUC_________UAAAGCCAUAAAAUUGGCAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGGCGCGAAACAUU .........((......((((((..........))))))..............(((((...((((((....))))))...)))))..................))........ (-17.96 = -17.68 + -0.28)
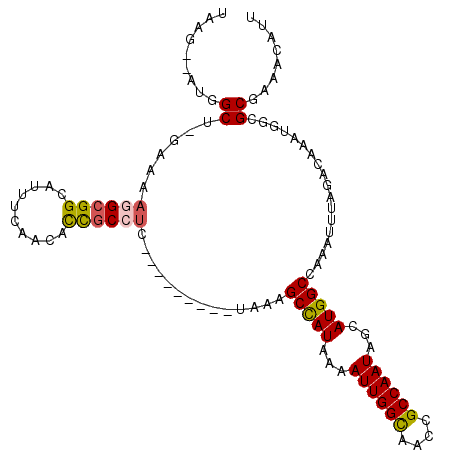
| Location | 41,324 – 41,426 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.80 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -21.39 |
| Energy contribution | -20.75 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
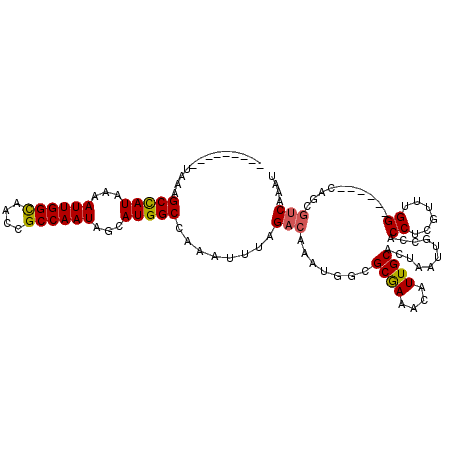
>2L_DroMel_CAF1 41324 102 - 22407834 AAUGAUUUGCGCCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUACCAAUUUAAUGGCUUUA---------GAGGCGAUGUUGAAAUGCCGCUUUUUCAAGCCAU--CUUA ..........(((.....(((((....((((((...(((((......)))))...)))))))))---------)))))((((((((((........))))))..)))--)... ( -26.30) >DroPse_CAF1 21981 106 - 1 AAUGUUUCGCGUCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUGCCAAUUUGAUGGCUUUAAAUAAGUCGGAGGCAGUGUUGAAAUGCCGCCUC-----GCCAA--CUUA ...(((..((...((((((.(((....((((((...((((((....))))))...))))))))).))))))..(((((.((((....)))).)))))-----)).))--)... ( -29.90) >DroEre_CAF1 18920 102 - 1 AAUGGUUCGCGCCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUACCAAUUUUAUAGCUUCA---------GAGGCGGUGUUGAAAUGCCGCUUUUUCAAGCAAU--AAGA ..((((((((((((..(......)..))))((......))))))..)))).((((((.((((.(---------((((((((((....)))))))))))..)))).))--)))) ( -32.40) >DroYak_CAF1 18183 102 - 1 AAUGGUUCGCGCAAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUACCAAUUUUAUAGCUUCA---------GGGGCGAUGUUGAAAUGCCUCCUUUUCCAGCCAU--CUUA .((((((.(((((....(((.......)))..))).(((((......)))))......))...(---------((((((.(.....).)))).))).....))))))--.... ( -20.30) >DroAna_CAF1 20662 113 - 1 AAUGUUUCGCGCCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUGCCAAUUUUAUGGCCUCAAAUAAGUCGGAGGCAGUGUUGAAAUGCCGCCUAGGCUGGCCAAGUCUGG ...................(((.(((((((((.(((...((((((...((((......(((((..........)))))...))))....))))))..)))))))))))).))) ( -40.00) >DroPer_CAF1 21741 106 - 1 AAUGUUUCGCGUCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUGCCAAUUUGAUGGCUUUAAAUAAGUCGGAGGCAGUGUUGAAAUGCCGCCUC-----GCCAA--CUUA ...(((..((...((((((.(((....((((((...((((((....))))))...))))))))).))))))..(((((.((((....)))).)))))-----)).))--)... ( -29.90) >consensus AAUGUUUCGCGCCAUUUGUCUAAAUUUGGCCAUGCUAUUGGCGGUUACCAAUUUUAUGGCUUCA_________GAGGCAGUGUUGAAAUGCCGCCUUUUC_AGCCAA__CUUA ........((((((..(......)..)))).........((((((...((((......(((((..........)))))...))))....)))))).......))......... (-21.39 = -20.75 + -0.64)
| Location | 41,362 – 41,460 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -18.31 |
| Energy contribution | -18.03 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
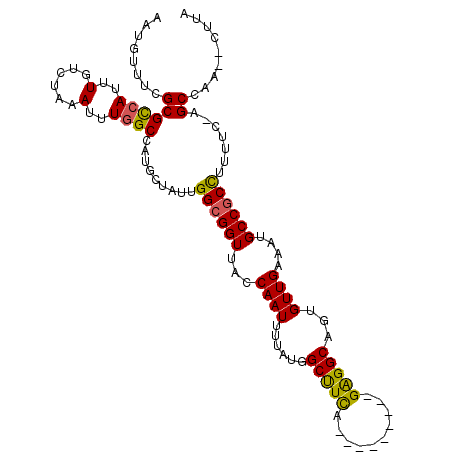
>2L_DroMel_CAF1 41362 98 + 22407834 ---------UAAAGCCAUUAAAUUGGUAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGGCGCAAAUCAUUGCACUAAUUGCCACCUCGUUUGG------CAGCGUCAAAU ---------....(((((...((((((....))))))...)))))........(((...(((.((((....)))).))).((((((.......)))------))).))).... ( -27.30) >DroPse_CAF1 22014 113 + 1 CGACUUAUUUAAAGCCAUCAAAUUGGCAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGACGCGAAACAUUGCACUAAUUGCCACCUCGUUUGGUGGCAACAUCGUCGAAU ((((...(((((((((((...((((((....))))))...)))))....))))))...(((..((((....)))).....((((((((......))))))))))).))))... ( -36.50) >DroEre_CAF1 18958 98 + 1 ---------UGAAGCUAUAAAAUUGGUAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGGCGCGAACCAUUGCACUAAUUGCCACCUCGUUCGG------CACCGUCAACC ---------....(((((...((((((....))))))...)))))........(((...(((.((((....)))).)))..((((.........))------))..))).... ( -22.70) >DroYak_CAF1 18221 98 + 1 ---------UGAAGCUAUAAAAUUGGUAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUUGCGCGAACCAUUGCACUAAUUGCCACCUCGUUCGG------CAGCGUCAAAC ---------....(((((...((((((....))))))...)))))........(((...((((.(((((....(((.....)))......))))))------))).))).... ( -23.00) >DroAna_CAF1 20702 105 + 1 CGACUUAUUUGAGGCCAUAAAAUUGGCAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGGCGCGAAACAUUGCACUAAUUGCCACCUCGUUUGG------CAGCCGC--AU ............((((((...((((((....))))))...))))))..............(((((((....))))......(((((.......)))------)))))..--.. ( -32.60) >DroPer_CAF1 21774 113 + 1 CGACUUAUUUAAAGCCAUCAAAUUGGCAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGACGCGAAACAUUGCACUAAUUGCCACCUCGUUUGGUGGCAACAUCGUCGAAU ((((...(((((((((((...((((((....))))))...)))))....))))))...(((..((((....)))).....((((((((......))))))))))).))))... ( -36.50) >consensus _________UAAAGCCAUAAAAUUGGCAACCGCCAAUAGCAUGGCCAAAUUUAGACAAAUGGCGCGAAACAUUGCACUAAUUGCCACCUCGUUUGG______CAGCGUCAAAU .............(((((...((((((....))))))...)))))........(((.......((((....))))...........((......))..........))).... (-18.31 = -18.03 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:11 2006