Index >
Results for CNB 481656_2
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 C-GCCCTGCCCATGTTTTGTGTGTCTCTGTCCATCCAGACTCCTGACGTTCAAGTTCGCA
481656_ENSG00000119866_HUMAN_9496_9947/1-454 C-GCCCTGCCCATGTTTTGTGTGTCTCTGTCCATCCAGACTCCTGACGTTCAAGTTCGCA
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 C-GCCCTGCCCATGTTTTGTGTGTCTCTGTCCATCCAGACTCCTGACGTTCAAGTTCGCA
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 TTGCCCTGCCCATGTTCTGTATGTCTGTGTCCATCCAT-CTCCTGACGTTCAAGTTCGCA
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 GGGACGTCACGTCCGCACTTGAACTTGCAGCTCAGGGGGGCTTTTGCCATTTTTTTCANN
481656_ENSG00000119866_HUMAN_9496_9947/1-454 GGGACGTCACGTCCGCACTTGAACTTGCAGCTCAGGGGGGCTTTTGCCATTTTTTTCANN
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 GGGACGTCACGTCCGCACTTGAACTTGCAGCTCAGGGGGGCTTTTGCCATTTTTTTCANN
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 GGGACGTCACGTCCGCACTTGAACTTGCAGCTCAGGGGGGCTTTTGCCATTTTTT-CATC
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTTTTTTTTTC
481656_ENSG00000119866_HUMAN_9496_9947/1-454 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTT------TTTTTCCTTN
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 TC------------------------------------------------TCTCTCTCTC
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 TTNNNNNNNNNNNNNN----------GCCATGACGGCTCTCCCACAATTCATCTTCCCTG
481656_ENSG00000119866_HUMAN_9496_9947/1-454 NNNNNNNNNNNNNNGCTTAAAAAAAAGCCATGACGGCTCTCCCACAATTCATCTTCCCTG
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 NNNNNNNNNNN---------------GCCATGACGGCTCTCCCACAATTCATCTTCCCTG
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 TTTTCAAAAAA---------------GCCATGACGGCTATCCCACAATCCATCTTCCCTG
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 CGCCATCTTTGTATTATTTCTAATTTATTTTGGATGTCAAAAGGCACTGATGAAGATATT
481656_ENSG00000119866_HUMAN_9496_9947/1-454 CGCCATCTTTGTATTATTTCTAATTTATTTTGGATGTCAAAAGGCACTGATGAAGATATT
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 CGCCATCTTTGTATTATTTCTAATTTATTTTGGATGTCAAAAGGCACTGATGAAGATATT
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 CGCCATCTTTGTATTATTTCTAATTTATTTTGGATGTCAAA-GGCATTGATGAAGATATT
481656_2.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 300
Mean pairwise identity: 81.21
Mean single sequence MFE: -58.51
Consensus MFE: -54.49
Energy contribution: -54.17
Covariance contribution: -0.31
Mean z-score: -1.71
Structure conservation index: 0.93
SVM decision value: 1.22
SVM RNA-class probability: 0.932280
Prediction: RNA
######################################################################
>481656_ENSRNOG00000007049_RAT_9511_9952/1-454
CGCCCUGCCCAUGUUUUGUGUGUCUCUGUCCAUCCAGACUCCUGACGUUCAAGUUCGCAGGGACGUCACGUCCGCACUUGAACUUGCAGCUCAGGGGGGCUUUUGCCAUUUUUUUCANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUUUUUUUUUCUUNNNNNNNNNNNNNNGCCAUGACGGCUCUCCCACAAUUCAUCUUCCCUGCGCCAUCUUUGUAUUAUUUCUAAUUUAUUUUGGAUGUCAAAAGGCACUGAUGAAGAUAUU
(((.......(((..(((((.(...(((((........(((((((.(((((((((((..((((((...))))).)...)))))))).))))))))))(((....))).............................................................................................)))))....).)))))..))).......)))..(((((..(......(((((......)))))((((....))))...)..)))))... ( -58.34)
>481656_ENSG00000119866_HUMAN_9496_9947/1-454
CGCCCUGCCCAUGUUUUGUGUGUCUCUGUCCAUCCAGACUCCUGACGUUCAAGUUCGCAGGGACGUCACGUCCGCACUUGAACUUGCAGCUCAGGGGGGCUUUUGCCAUUUUUUUCANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGCUUAAAAAAAAGCCAUGACGGCUCUCCCACAAUUCAUCUUCCCUGCGCCAUCUUUGUAUUAUUUCUAAUUUAUUUUGGAUGUCAAAAGGCACUGAUGAAGAUAUU
(((.......(((..(((((.(...(((((......(((((((((.(((((((((((..((((((...))))).)...)))))))).))))))))))(((....))).......)).............................................................................((((......))))...)))))....).)))))..))).......)))..(((((..(......(((((......)))))((((....))))...)..)))))... ( -60.54)
>481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454
CGCCCUGCCCAUGUUUUGUGUGUCUCUGUCCAUCCAGACUCCUGACGUUCAAGUUCGCAGGGACGUCACGUCCGCACUUGAACUUGCAGCUCAGGGGGGCUUUUGCCAUUUUUUUCANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUUUUUUUCCUUNNNNNNNNNNNNGCCAUGACGGCUCUCCCACAAUUCAUCUUCCCUGCGCCAUCUUUGUAUUAUUUCUAAUUUAUUUUGGAUGUCAAAAGGCACUGAUGAAGAUAUU
(((.......(((..(((((.(...(((((........(((((((.(((((((((((..((((((...))))).)...)))))))).))))))))))(((....)))..................................................................................)))))....).)))))..))).......)))..(((((..(......(((((......)))))((((....))))...)..)))))... ( -58.34)
>481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454
UUGCCCUGCCCAUGUUCUGUAUGUCUGUGUCCAUCCAUCUCCUGACGUUCAAGUUCGCAGGGACGUCACGUCCGCACUUGAACUUGCAGCUCAGGGGGGCUUUUGCCAUUUUUUCAUCUCUCUCUCUCUCUUUUCAAAAAAGCCAUGACGGCUAUCCCACAAUCCAUCUUCCCUGCGCCAUCUUUGUAUUAUUUCUAAUUUAUUUUGGAUGUCAAAGGCAUUGAUGAAGAUAUU
...........((.(((...((((((.((..((((((.........((((((((..((..(((((...))))))))))))))).....((.(((((((((....))).................................((((.....))))................)))))).))...........................)))))).)).))))))....))).))... ( -56.80)
>consensus
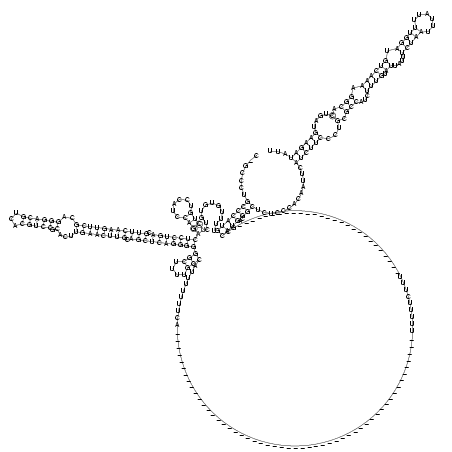
C_GCCCUGCCCAUGUUUUGUGUGUCUCUGUCCAUCCAGACUCCUGACGUUCAAGUUCGCAGGGACGUCACGUCCGCACUUGAACUUGCAGCUCAGGGGGGCUUUUGCCAUUUUUUUCA____________________________________________________UUUUUCUUU___________________________GCCAUGACGGCUCUCCCACAAUUCAUCUUCCCUGCGCCAUCUUUGUAUUAUUUCUAAUUUAUUUUGGAUGUCAAAAGGCACUGAUGAAGAUAUU
.......(((((((...........((((......))))(((((((.(((((((((((..((((((...))))).)...)))))))).))))))))))(((....)))....................................................................................................))))..))).............((((((...(.(((...((((.((....(((((......))))).)))))).))).)....))))))... (-54.49 = -54.17 + -0.31)
481656_2.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004