Index >
Results for CNB 478948-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
478948_ENSMUSG00000026565_MOUSE_4444_4538/1-97 -TGTCCGAGGAAGGCCTGGGCCTGGGCTGTGGAGATCGCTCCTCCAACGGGCACGGGCTG
478948_SINFRUG00000130458_FUGU_7759_7854/1-97 -TGTTGGAGGAGGGCTTGTGCATGTGCATTAGTGATTGCACTTGTGGTTGGCACCATTTG
478948_ENSRNOG00000003581_RAT_3735_3829/1-97 -TGCCCAAGGAAAGCCTGGGCCTGGGCTGTGGAGATGGCCCCTCCGACAGGCACGGGCTG
478948_ENSDARG00000007996_ZEBRAFISH_8031_8126/1-97 CTGTTGGAGCAAGGCCTGTGCGTGTGCGTTGGTGATTGCGCTTGTTGTTTGCACAGTCTG
** ** * ** ** ** ** ** * * *** ** * * **** **
478948_ENSMUSG00000026565_MOUSE_4444_4538/1-97 TTTCTGAAAGTCCAGACCATTGGTTTGTGTGCCTGG-
478948_SINFRUG00000130458_FUGU_7759_7854/1-97 CCTCTGGAAATCCAATCCATTTATTTGGATACCTGGG
478948_ENSRNOG00000003581_RAT_3735_3829/1-97 CTTCTCAAAGTCCAGGCCATTGGTCTGTGTGCCTGG-
478948_ENSDARG00000007996_ZEBRAFISH_8031_8126/1-97 CCTCTGAAAGTCCAGTCCATTTGTTTGGGCACCTGG-
*** ** **** ***** * ** *****
478948-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 97
Mean pairwise identity: 70.83
Mean single sequence MFE: -36.17
Consensus MFE: -23.59
Energy contribution: -24.78
Covariance contribution: 1.19
Mean z-score: -1.33
Structure conservation index: 0.65
SVM decision value: 0.17
SVM RNA-class probability: 0.618566
Prediction: RNA
######################################################################
>478948_ENSMUSG00000026565_MOUSE_4444_4538/1-97
UGUCCGAGGAAGGCCUGGGCCUGGGCUGUGGAGAUCGCUCCUCCAACGGGCACGGGCUGUUUCUGAAAGUCCAGACCAUUGGUUUGUGUGCCUGG
.((((.....((((....))))))))..(((((.......))))).((((((((((((.........)))))(((((...)))))..))))))). ( -38.10)
>478948_SINFRUG00000130458_FUGU_7759_7854/1-97
UGUUGGAGGAGGGCUUGUGCAUGUGCAUUAGUGAUUGCACUUGUGGUUGGCACCAUUUGCCUCUGGAAAUCCAAUCCAUUUAUUUGGAUACCUGGG
..(..((((.(((((..((((.(((((........))))).))))...))).)).....))))..)....((((((((......)))))...))). ( -29.90)
>478948_ENSRNOG00000003581_RAT_3735_3829/1-97
UGCCCAAGGAAAGCCUGGGCCUGGGCUGUGGAGAUGGCCCCUCCGACAGGCACGGGCUGCUUCUCAAAGUCCAGGCCAUUGGUCUGUGUGCCUGG
...(((.((..((((..((((((((((...((((.(((((..((....))...)))))...))))..))))))))))...)).)).....))))) ( -45.60)
>478948_ENSDARG00000007996_ZEBRAFISH_8031_8126/1-97
CUGUUGGAGCAAGGCCUGUGCGUGUGCGUUGGUGAUUGCGCUUGUUGUUUGCACAGUCUGCCUCUGAAAGUCCAGUCCAUUUGUUUGGGCACCUGG
....((((.(((((((((((((...(((..((((....))))...))).)))))))...)))).))....))))(((((......)))))...... ( -31.10)
>consensus
_UGUCCGAGGAAGGCCUGGGCCUGGGCUGUGGAGAUUGCACCUCCGACUGGCACGGGCUGCCUCUGAAAGUCCAGUCCAUUGGUUUGGGUACCUGG_
...(((((((..((((((((((((......((((....)))).....)))))))))))).)))))))..((((((((.....))))))))....... (-23.59 = -24.78 + 1.19)
478948-rev.rnaz
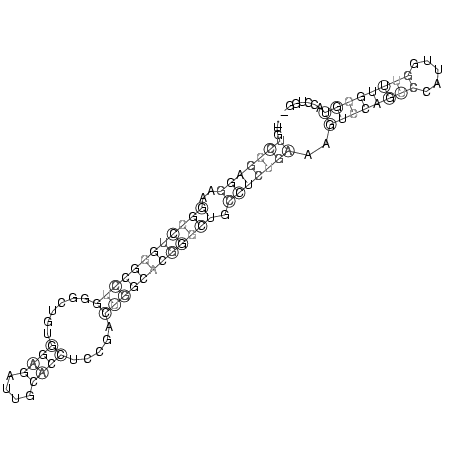
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004