Index >
Results for CNB 461739
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
461739_ENSMUSG00000034255_MOUSE_8785_8894/1-112 -CCCTCCAGCTTCGAAGCCGAGATGGCTC-GGAGTACCTGATCCAGCACGACTCCGAAGC
461739_SINFRUG00000131017_FUGU_9484_9593/1-112 CCTCCCCAG-TTAAAAACTAAGAACGCGGTGGAATTCCTGATACAGTACGACACAGAAAG
461739_ENSRNOG00000028569_RAT_8775_8884/1-112 -CCCTCCAGCTCCGGAGCCGAGATGGCTC-AGAGTACCTGATCCAGCATGACTCAGAGGC
461739_ENSDARG00000006344_ZEBRAFISH_6913_7022/1-112 CTCATTCAGCTTAAGACTCGGACTGGTGC-CGAATACTTGATCCAGTACGACACCGATAG
*** * * * ** * * **** *** * *** * **
461739_ENSMUSG00000034255_MOUSE_8785_8894/1-112 CATCATCAGCACCTGGCACAAAGCCATTGCCGAAGGCATCTCAGAGCTGGTA
461739_SINFRUG00000131017_FUGU_9484_9593/1-112 CATCATTGTTGACTGGCACCGAGTCCTGACCGATACCATACGACAACTGGT-
461739_ENSRNOG00000028569_RAT_8775_8884/1-112 CATCATCAGCACCTGGCACAAGGCCATTGCCGAAGGCATCGAGGAGCTGGTA
461739_ENSDARG00000006344_ZEBRAFISH_6913_7022/1-112 CATCATCCAAGACTGGCATAAAGTTATCTTAGATACCATACACCAGCTGGT-
****** ****** * * ** *** * *****
//
461739.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 112
Mean pairwise identity: 65.37
Mean single sequence MFE: -36.25
Consensus MFE: -16.50
Energy contribution: -18.75
Covariance contribution: 2.25
Mean z-score: -2.29
Structure conservation index: 0.46
SVM decision value: 1.60
SVM RNA-class probability: 0.966849
Prediction: RNA
######################################################################
>461739_ENSMUSG00000034255_MOUSE_8785_8894/1-112
CCCUCCAGCUUCGAAGCCGAGAUGGCUCGGAGUACCUGAUCCAGCACGACUCCGAAGCCAUCAUCAGCACCUGGCACAAAGCCAUUGCCGAAGGCAUCUCAGAGCUGGUA
....((((((..((.(((..(((((((((((((..(((...)))....)))))).))))))).((.(((..((((.....)))).))).)).)))...))..)))))).. ( -49.40)
>461739_SINFRUG00000131017_FUGU_9484_9593/1-112
CCUCCCCAGUUAAAAACUAAGAACGCGGUGGAAUUCCUGAUACAGUACGACACAGAAAGCAUCAUUGUUGACUGGCACCGAGUCCUGACCGAUACCAUACGACAACUGGU
.....((((((........((.((.((((((....)).....((((.(((((..((.....))..)))))))))..)))).)).))...((........))..)))))). ( -23.90)
>461739_ENSRNOG00000028569_RAT_8775_8884/1-112
CCCUCCAGCUCCGGAGCCGAGAUGGCUCAGAGUACCUGAUCCAGCAUGACUCAGAGGCCAUCAUCAGCACCUGGCACAAGGCCAUUGCCGAAGGCAUCGAGGAGCUGGUA
....((((((((.(((((..((((((((.((((..(((...)))....)))).)).)))))).((.(((..((((.....)))).))).)).))).))..)))))))).. ( -48.70)
>461739_ENSDARG00000006344_ZEBRAFISH_6913_7022/1-112
CUCAUUCAGCUUAAGACUCGGACUGGUGCCGAAUACUUGAUCCAGUACGACACCGAUAGCAUCAUCCAAGACUGGCAUAAAGUUAUCUUAGAUACCAUACACCAGCUGGU
.....((((((......(((.(((((...(((....)))..))))).)))....((((((...(((((....))).))...))))))................)))))). ( -23.00)
>consensus
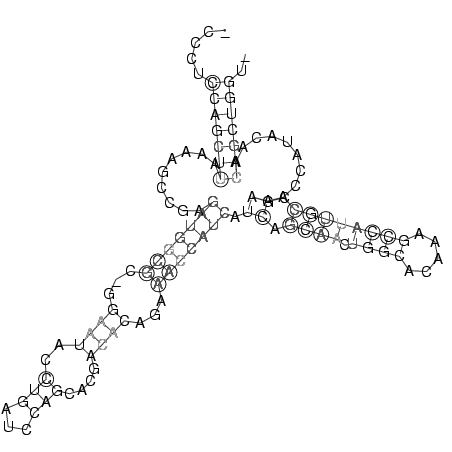
_CCCUCCAGCUUAAAAGCCGAGAUGGCGC_GGAAUACCUGAUCCAGCACGACACAGAAACCAUCAUCAGCAACUGGCACAAAGCCAUUGCCGAAACCAUACAACAGCUGGU_
.....(((((((.........(((((((...((((..(((...)))....))))...))))))).((.((((.((((.....)))))))).))..........))))))).. (-16.50 = -18.75 + 2.25)
461739.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004