Index >
Results for CNB 414303
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
414303_ENSG00000143549_HUMAN_1805_1947/1-143 TAGCTGGAGGATGAGCTGGCAGCCATGCAGAAGAAGCTGAAAGGGACAGAGGATGAGCTG
414303_SINFRUG00000130484_FUGU_7340_7481/1-143 -AGCTTGAGGACGATTTGGTAGCCCTGCAGAAGAAATTAAAGGGGACAGAAGATGAGTTG
414303_ENSMUSG00000027940_MOUSE_3986_4126/1-143 -AGCTAGAGGATGAACTAGCAACCATGCAGAAGAAGCTGAAAGGGACAGAGGATGAGCTG
414303_ENSRNOG00000017441_RAT_4521_4661/1-143 -AGCTAGAGGATGAACTAGCAACCATGCAGAAGAAGCTGAAAGGGACAGAGGATGAGCTG
**** ***** ** * * * ** ********** * ** ******** ****** **
414303_ENSG00000143549_HUMAN_1805_1947/1-143 GACAAGTATTCTGAAGCTTTGAAGGATGCCCAGGAGAAGCTGGAACTGGCAGAGAAGAAG
414303_SINFRUG00000130484_FUGU_7340_7481/1-143 GACAAGTTCTCCGAGGCTCTTAAAGATGCTCAGGAGAAGCTTGAACTGGCAGAAAAGAAA
414303_ENSMUSG00000027940_MOUSE_3986_4126/1-143 GACAAGTATTCGGAAGCTTTAAAGGATGCTCAGGAGAAGCTGGAGCTAGCAGAGAAGAAG
414303_ENSRNOG00000017441_RAT_4521_4661/1-143 GACAAGTATTCTGAGGCTTTAAAGGATGCCCAGGAGAAGCTGGAGCTAGCAGAGAAGAAG
******* ** ** *** * ** ***** *********** ** ** ***** *****
414303_ENSG00000143549_HUMAN_1805_1947/1-143 GCTGCTGATGTGAGTATTAGAGA
414303_SINFRUG00000130484_FUGU_7340_7481/1-143 GCCACAGACGTAAGTATGGGGGA
414303_ENSMUSG00000027940_MOUSE_3986_4126/1-143 GCAGCCGACGTGAGTATTGGGG-
414303_ENSRNOG00000017441_RAT_4521_4661/1-143 GCAGCGGATGTGAGTATTGGGA-
** * ** ** ***** *
//
414303.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 143
Mean pairwise identity: 84.19
Mean single sequence MFE: -35.94
Consensus MFE: -24.88
Energy contribution: -24.50
Covariance contribution: -0.37
Mean z-score: -1.66
Structure conservation index: 0.69
SVM decision value: 0.21
SVM RNA-class probability: 0.638540
Prediction: RNA
######################################################################
>414303_ENSG00000143549_HUMAN_1805_1947/1-143
UAGCUGGAGGAUGAGCUGGCAGCCAUGCAGAAGAAGCUGAAAGGGACAGAGGAUGAGCUGGACAAGUAUUCUGAAGCUUUGAAGGAUGCCCAGGAGAAGCUGGAACUGGCAGAGAAGAAGGCUGCUGAUGUGAGUAUUAGAGA
.((((........))))(((((((...(((((...(((.....(..(((........)))..).))).)))))...(((((..((.(..((((......)))).)))..))))).....)))))))................. ( -37.90)
>414303_SINFRUG00000130484_FUGU_7340_7481/1-143
AGCUUGAGGACGAUUUGGUAGCCCUGCAGAAGAAAUUAAAGGGGACAGAAGAUGAGUUGGACAAGUUCUCCGAGGCUCUUAAAGAUGCUCAGGAGAAGCUUGAACUGGCAGAAAAGAAAGCCACAGACGUAAGUAUGGGGGA
.((((........((((....((((....((....))...)))).))))......((..(.((((((((((..(((((.....)).)))..)))).))))))..)..))........))))..((.((....)).))..... ( -36.80)
>414303_ENSMUSG00000027940_MOUSE_3986_4126/1-143
AGCUAGAGGAUGAACUAGCAACCAUGCAGAAGAAGCUGAAAGGGACAGAGGAUGAGCUGGACAAGUAUUCGGAAGCUUUAAAGGAUGCUCAGGAGAAGCUGGAGCUAGCAGAGAAGAAGGCAGCCGACGUGAGUAUUGGGG
.(((((........)))))..((.....((((...(((((..(..(((........)))..).....)))))...))))....((((((((.(....((((....)))).........((...))..).)))))))))).. ( -34.00)
>414303_ENSRNOG00000017441_RAT_4521_4661/1-143
AGCUAGAGGAUGAACUAGCAACCAUGCAGAAGAAGCUGAAAGGGACAGAGGAUGAGCUGGACAAGUAUUCUGAGGCUUUAAAGGAUGCCCAGGAGAAGCUGGAGCUAGCAGAGAAGAAGGCAGCGGAUGUGAGUAUUGGGA
.(((((........)))))..((((((.......((((.......((.....)).(((((...(((.(((((.(((..........))))))))...)))....)))))...........))))........))).))).. ( -35.06)
>consensus
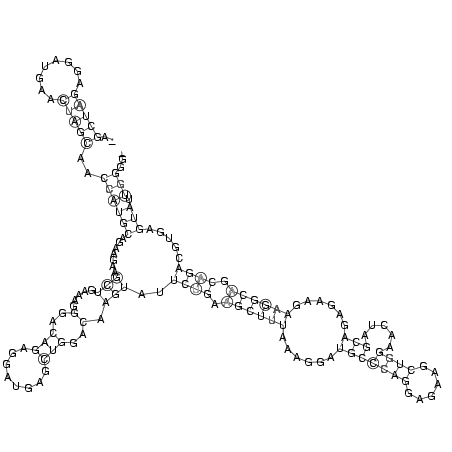
_AGCUAGAGGAUGAACUAGCAACCAUGCAGAAGAAGCUGAAAGGGACAGAGGAUGAGCUGGACAAGUAUUCUGAAGCUUUAAAGGAUGCCCAGGAGAAGCUGGAACUAGCAGAGAAGAAGGCAGCAGACGUGAGUAUUGGGG_
..(((((........)))))..((((((.......(((.....(..(((........)))..).)))..((((.((((((......(((((((......)))).....)))......)))))).)))).....))).)))... (-24.88 = -24.50 + -0.37)
414303.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004