Index >
Results for CNB 391318
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
391318_ENSG00000180806_HUMAN_1484_1578/1-96 CTTAGGTAGTTTCATGTTGTTGGGATTGAGTTTTGAACTCGGCAACAAGAAACTGCCTGA
391318_ENSDARG00000013621_ZEBRAFISH_654_741/1-96 GAGTGGGAGACCCATAACGTTG--ATTTAAATATTATCCAGGTGACCACAGTAAGTCAAG
391318_ENSMUSG00000036139_MOUSE_1625_1719/1-96 CTTAGGTAGTTTCATGTTGTTGGGATTGAGTTTTGAACTCGGCAACAAGAAACTGCCTGA
391318_SINFRUG00000146331_FUGU_536_621/1-96 -AGTGAAAGACC-ATAACGTTG--ATTTAAATATTATCCAGGTGACCACAGTAAGTCAAG
* ** ** **** *** * * * * * ** ** * * * *
391318_ENSG00000180806_HUMAN_1484_1578/1-96 GTTACATCAGTCGGTTTTCGTCGAGGGCCCCAACC-
391318_ENSDARG00000013621_ZEBRAFISH_654_741/1-96 GTCATAAAATTGTAATGTCATGACGGTCC------T
391318_ENSMUSG00000036139_MOUSE_1625_1719/1-96 GTTACATCAGTCGGTTTTCGTCGAGGGCCCCAATC-
391318_SINFRUG00000146331_FUGU_536_621/1-96 GTCATAAAATTCTAATGTCAGGTTCGCCC------T
** * * * * * ** * **
//
391318.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 96
Mean pairwise identity: 58.20
Mean single sequence MFE: -26.13
Consensus MFE: -10.96
Energy contribution: -16.28
Covariance contribution: 5.31
Mean z-score: -1.91
Structure conservation index: 0.42
SVM decision value: 0.68
SVM RNA-class probability: 0.820585
Prediction: RNA
######################################################################
>391318_ENSG00000180806_HUMAN_1484_1578/1-96
CUUAGGUAGUUUCAUGUUGUUGGGAUUGAGUUUUGAACUCGGCAACAAGAAACUGCCUGAGUUACAUCAGUCGGUUUUCGUCGAGGGCCCCAACC
(((((((((((((.((((((((((.(..(...)..).)))))))))).)))))))))))))...........(((((((...)))))))...... ( -37.40)
>391318_ENSDARG00000013621_ZEBRAFISH_654_741/1-96
GAGUGGGAGACCCAUAACGUUGAUUUAAAUAUUAUCCAGGUGACCACAGUAAGUCAAGGUCAUAAAAUUGUAAUGUCAUGACGGUCCU
..(((((...)))))..((((.......(((((((....((((((............))))))......)))))))...))))..... ( -16.20)
>391318_ENSMUSG00000036139_MOUSE_1625_1719/1-96
CUUAGGUAGUUUCAUGUUGUUGGGAUUGAGUUUUGAACUCGGCAACAAGAAACUGCCUGAGUUACAUCAGUCGGUUUUCGUCGAGGGCCCCAAUC
(((((((((((((.((((((((((.(..(...)..).)))))))))).)))))))))))))...........(((((((...)))))))...... ( -37.40)
>391318_SINFRUG00000146331_FUGU_536_621/1-96
AGUGAAAGACCAUAACGUUGAUUUAAAUAUUAUCCAGGUGACCACAGUAAGUCAAGGUCAUAAAAUUCUAAUGUCAGGUUCGCCCU
.(((((.(((.((((..((......))..))))....((((((............))))))...........)))...)))))... ( -13.50)
>consensus
CAGAGGUAGACCCAUAACGUUG__AUUGAAAUAUGAACCAGGCAACAACAAAAAGCCAAAGUCACAAAAGUCGAAUGUCAUCGAGGGCC_______
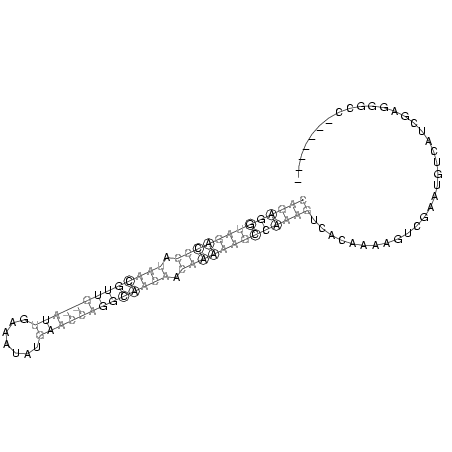
(((((((((((((.(((((((((((((.......))))))))))))).)))))))))))))................................... (-10.96 = -16.28 + 5.31)
391318.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004