Index >
Results for CNB 244427_1-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
244427_ENSMUSG00000036888_MOUSE_6983_7489/1-510 TGAGAAGGGCTGCGGAAGATAATTTATAGGTTTTAATTACTCTTCATTCCGCCTGATCAG
244427_ENSRNOG00000002457_RAT_6069_6575/1-510 TGAGAAGGGCTGCGGAAGATAATTTATAGGTTTTAATTACTCTTCATTCCGCCTGATCAG
244427_ENSDARG00000015073_ZEBRAFISH_4427_4928/1-510 TGAGAGGGACTGCAAAAGATAATTTATAGGTTTTAATTAGCGCCCATTCCGCCTGATCAG
244427_ENSG00000163064_HUMAN_6937_7444/1-510 CGAGAAGGGCTGCGGAAGATAATTTATAGGTTTTAATTACTCTTCATTCCGCCTGATCAG
**** ** **** ************************ ****************
244427_ENSMUSG00000036888_MOUSE_6983_7489/1-510 CTTGGCGTTCATCACAGGCCTGTGAATAAAACCCTGGTCAAAAGCTCTGTCACATAGCAC
244427_ENSRNOG00000002457_RAT_6069_6575/1-510 CTTGGCGTTCATCACAGGCCTGTGAATAAAACCCTGGTCAAAAGCTCTGTCACATAGCGC
244427_ENSDARG00000015073_ZEBRAFISH_4427_4928/1-510 CTTGGCGTTCATCACTAGCGAGTGAATAAAAGCC-GGTCAAAAGCACTGTCACTACCCGC
244427_ENSG00000163064_HUMAN_6937_7444/1-510 CTTGGCGTTCATCACAGGCCTGTGAATAAAACCCTGGTCAAAAGCTCTGTCACATCGCGC
*************** ** ********** ** ********** ******* * *
244427_ENSMUSG00000036888_MOUSE_6983_7489/1-510 TGGCAAGACGTTTAATCAAAGTGA-NNNNNNNNNNNNNNNNNNNNNNNNNNTCCTGTACG
244427_ENSRNOG00000002457_RAT_6069_6575/1-510 TGGCAAGACGTTTAATCAAAGTGA-NNNNNNNNNNNNNNNNNNNNNNNNNNTCCTGTACG
244427_ENSDARG00000015073_ZEBRAFISH_4427_4928/1-510 TGGCAGGGCGCTTTATCAAGGTAAGCAGGGCCGTATTACACGAGGTGTAGA--GGCAACG
244427_ENSG00000163064_HUMAN_6937_7444/1-510 TGGCAAGACGCTTAATCAAAGTGANNNNNNNNNNNNNNNNNNNNNNNNNNNTCCTCCACG
***** * ** ** ***** ** * ***
244427_ENSMUSG00000036888_MOUSE_6983_7489/1-510 TAATTTCCAGCCAGCTGATAAAGCCAGCTGAATAATGCGGCGGCCCTTTGGAGACACCAT
244427_ENSRNOG00000002457_RAT_6069_6575/1-510 TAATTTCCAGCCAGCTGATAAAGCCAGCTGAATAATGCGGCGGCCCTTTGGAGACACCAT
244427_ENSDARG00000015073_ZEBRAFISH_4427_4928/1-510 TAATTTCCCCAGTGCTGATAAAGCCCGTTGAATAATGCCGGAGTCATCT-GAGACACCAT
244427_ENSG00000163064_HUMAN_6937_7444/1-510 TAATTTCCAGCCAGCTGATAAAGCCAGCTGAATAATACGGCGGCCCTTTGGAGACACCAT
******** ************ * ******** * * * * * * **********
244427_ENSMUSG00000036888_MOUSE_6983_7489/1-510 TTACAGAAATGACTTTATTGACGGCTTAACGTCGGTAATTCATTTTACCTTTCATGTAGT
244427_ENSRNOG00000002457_RAT_6069_6575/1-510 TTACAGAAATGACTTTATTGACGGCTTAACGTCGGTAATTCATTTTACCTTTCATGTAGT
244427_ENSDARG00000015073_ZEBRAFISH_4427_4928/1-510 TTACTGAAATGACTTTATTGACGACTTAACGTCGGTAATTCATTTT-CCCTTCACGTATC
244427_ENSG00000163064_HUMAN_6937_7444/1-510 TTACAGAAATGACTTTATTGACGGCTTAACGTCGGTAATTCATTTTACCTTTCATGTAGT
**** ****************** ********************** ** **** ***
244427_1-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 300
Mean pairwise identity: 85.77
Mean single sequence MFE: -80.82
Consensus MFE: -61.47
Energy contribution: -61.97
Covariance contribution: 0.50
Mean z-score: -1.38
Structure conservation index: 0.76
SVM decision value: -0.06
SVM RNA-class probability: 0.505275
Prediction: RNA
######################################################################
>244427_ENSMUSG00000036888_MOUSE_6983_7489/1-510
UGAGAAGGGCUGCGGAAGAUAAUUUAUAGGUUUUAAUUACUCUUCAUUCCGCCUGAUCAGCUUGGCGUUCAUCACAGGCCUGUGAAUAAAACCCUGGUCAAAAGCUCUGUCACAUAGCACUGGCAAGACGUUUAAUCAAAGUGANNNNNNNNNNNNNNNNNNNNNNNNNNUCCUGUACGUAAUUUCCAGCCAGCUGAUAAAGCCAGCUGAAUAAUGCGGCGGCCCUUUGGAGACACCAUUUACAGAAAUGACUUUAUUGACGGCUUAACGUCGGUAAUUCAUUUUACCUUUCAUGUAGU
...((((((((((.((((((((((..........))))).)))))....(((....((((((.(((..((((((((....)))))........((((......(((.((...)).))).((((....((((.............................................)))).....)))))))).)))....))))))))).....)))))))))))))(....).....((((((((((((..(((((((((......))))))))).)))))))........))))). ( -77.78)
>244427_ENSRNOG00000002457_RAT_6069_6575/1-510
UGAGAAGGGCUGCGGAAGAUAAUUUAUAGGUUUUAAUUACUCUUCAUUCCGCCUGAUCAGCUUGGCGUUCAUCACAGGCCUGUGAAUAAAACCCUGGUCAAAAGCUCUGUCACAUAGCGCUGGCAAGACGUUUAAUCAAAGUGANNNNNNNNNNNNNNNNNNNNNNNNNNUCCUGUACGUAAUUUCCAGCCAGCUGAUAAAGCCAGCUGAAUAAUGCGGCGGCCCUUUGGAGACACCAUUUACAGAAAUGACUUUAUUGACGGCUUAACGUCGGUAAUUCAUUUUACCUUUCAUGUAGU
...((((((((((.((((((((((..........))))).)))))....(((....((((((.(((.....(((((....)))))...........((((...(((.((...)).)))((((((..((.(((.......((.((..........................))))......))).))..))))))))))...))))))))).....)))))))))))))(....).....((((((((((((..(((((((((......))))))))).)))))))........))))). ( -80.59)
>244427_ENSDARG00000015073_ZEBRAFISH_4427_4928/1-510
UGAGAGGGACUGCAAAAGAUAAUUUAUAGGUUUUAAUUAGCGCCCAUUCCGCCUGAUCAGCUUGGCGUUCAUCACUAGCGAGUGAAUAAAAGCCGGUCAAAAGCACUGUCACUACCCGCUGGCAGGGCGCUUUAUCAAGGUAAGCAGGGCCGUAUUACACGAGGUGUAGAGGCAACGUAAUUUCCCCAGUGCUGAUAAAGCCCGUUGAAUAAUGCCGGAGUCAUCUGAGACACCAUUUACUGAAAUGACUUUAUUGACGACUUAACGUCGGUAAUUCAUUUUCCCUUCACGUAUC
(((.(((((...((..((((........(((.(((.(((((((((.....((.(((((.((((...(((((((......)).)))))..)))).)))))...))..(((((........)))))))))((((.((....)))))).((((..((((((((..((......(....).......))...))).)))))..))))))))).))).)))((.(((......))).))))))..))((((((..(((((((((......))))))))).))))))))))))))...... ( -85.12)
>244427_ENSG00000163064_HUMAN_6937_7444/1-510
CGAGAAGGGCUGCGGAAGAUAAUUUAUAGGUUUUAAUUACUCUUCAUUCCGCCUGAUCAGCUUGGCGUUCAUCACAGGCCUGUGAAUAAAACCCUGGUCAAAAGCUCUGUCACAUCGCGCUGGCAAGACGCUUAAUCAAAGUGANNNNNNNNNNNNNNNNNNNNNNNNNNNUCCUCCACGUAAUUUCCAGCCAGCUGAUAAAGCCAGCUGAAUAAUACGGCGGCCCUUUGGAGACACCAUUUACAGAAAUGACUUUAUUGACGGCUUAACGUCGGUAAUUCAUUUUACCUUUCAUGUAGU
...(((((((((((((.((.........(((((((..............((((..........))))....(((((....))))).))))))).((((...(((((((((((........)))).))).)))).)))).................................)).))).((((.....((((..(((.....)))..)))).....))))))))))))))(....).....((((((((((((..(((((((((......))))))))).)))))))........))))). ( -79.80)
>consensus
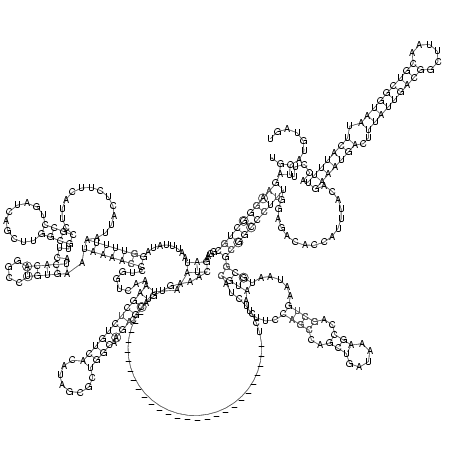
UGAGAAGGGCUGCGGAAGAUAAUUUAUAGGUUUUAAUUACUCUUCAUUCCGCCUGAUCAGCUUGGCGUUCAUCACAGGCCUGUGAAUAAAACCCUGGUCAAAAGCUCUGUCACAUAGCGCUGGCAAGACGCUUAAUCAAAGUGA___________________________UCCUCUACGUAAUUUCCAGCCAGCUGAUAAAGCCAGCUGAAUAAUGCGGCGGCCCUUUGGAGACACCAUUUACAGAAAUGACUUUAUUGACGGCUUAACGUCGGUAAUUCAUUUUACCUUUCAUGUAGU
.((((((((((((....(((........(((((((..............((((..........))))....(((((....))))).)))))))........(((((((((((........)))).))).)))).))).........................................((((.....((((..(((.....)))..)))).....))))))))))))))................(((((((..(((((((((......))))))))).))))))).....))....... (-61.47 = -61.97 + 0.50)
244427_1-rev.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004