Index >
Results for CNB 134270_43-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
134270_ENSDARG00000020961_ZEBRAFISH_5965_8474/1-2687 AGCACCCCACTGCATTTCAATGTTTAA-ATTTTA-ATGAACCCTTTAAAAATGTCTCAAG
134270_ENSG00000164853_HUMAN_5374_7839/1-2687 --C--CGCGCCGC-GGCGGCTCCGTGC-GTCCCA-ATG--C---T--CGCATTTC-AATC
134270_SINFRUG00000149320_FUGU_3315_5852/1-2687 AGCAGCCCTCTGTATTTCAGCGCGTGCAGCCCCACATT--C---TAACGAACTCCTCACA
134270_ENSRNOG00000001284_RAT_5040_7483/1-2687 --CCGTGCGCCGCAGCCGGCTCCACGC-ATCTCA-ATG--C---G--ACCATTTC-AATC
134270_ENSMUSG00000029546_MOUSE_5013_7498/1-2687 --CCGCGCGCTGCAGCCGGCTCCACGC-ATCTCA-ATG--C---G--ACCATTTC-AATC
* * * * * ** * * * *
134270_ENSDARG00000020961_ZEBRAFISH_5965_8474/1-2687 ATGTAGCCTTTTAT--CCCTCTATCTTACAAAAAGTGCTTTTAGAAGTTTG--GGAATCT
134270_ENSG00000164853_HUMAN_5374_7839/1-2687 GCCCTGTGTTTT-TCATGCTGTCGGGCCGGCT-CGCGCTCCGA-AAACCTCCTCGAGTGT
134270_SINFRUG00000149320_FUGU_3315_5852/1-2687 AATCTGCCTTAT-TCATCCTTCCTCCTCTTTTAAGCGCTT--C-TTTCTTC--GGATTT-
134270_ENSRNOG00000001284_RAT_5040_7483/1-2687 GCCCAGTGGCNN-N--NNNNNNNNNNNNNNNN-NNNNCTC--A--------------TG-
134270_ENSMUSG00000029546_MOUSE_5013_7498/1-2687 GCCCAGTGGCTT-T--TTNNNNNNNNNNNNNN-NNNNNNNNNN-NNNNNNNNNNNNNNNN
*
134270_ENSDARG00000020961_ZEBRAFISH_5965_8474/1-2687 TTGAATGTTTTTTTCTTTGTGTCTTAAACTTAT--TTTAT-----CATACTGTACACAAG
134270_ENSG00000164853_HUMAN_5374_7839/1-2687 TTGAGAGGGGCCTTTAATGACGGTGGGGGCGGCCGGGACGACCCCCGC--CCGGCTCGAG
134270_SINFRUG00000149320_FUGU_3315_5852/1-2687 -TGAGGGCGATCTG--ATGAATAATTGCCCCTC--TCTCG-----GTC--TCTTTTTGAA
134270_ENSRNOG00000001284_RAT_5040_7483/1-2687 -TGCTCGGGACCCT--NNNNNNNNNNNNNNNTC--TCAAG-----CGT--TGGAGTGGGG
134270_ENSMUSG00000029546_MOUSE_5013_7498/1-2687 GTGCCCGGGACCCTNNNNNNNNNNNNNNNNNNC--TCAAG-----CGT--TGGAGTGGGG
** *
134270_ENSDARG00000020961_ZEBRAFISH_5965_8474/1-2687 TATGAATAATTCGGTTTATCCAGATCCCCCTTTTGAACATCAGAATTCTATAATTGCAGC
134270_ENSG00000164853_HUMAN_5374_7839/1-2687 AGCTCTTCAGAGGCCCTCGGC-TGCGGCGGCGCCGCGGGGAGTCGCTCGGCTTCTGCTCT
134270_SINFRUG00000149320_FUGU_3315_5852/1-2687 CATCTAAAATTCAGTGATTGCTTTTGGTCATTTTCAGGCACGACTCCAACCNNNNNNNNN
134270_ENSRNOG00000001284_RAT_5040_7483/1-2687 GACTTTAATGACGGTGGGGGT-GGCCGAGTCGACCCCGCCCGGCTCGAGAGCTCTTCAGA
134270_ENSMUSG00000029546_MOUSE_5013_7498/1-2687 GACTTTAATGACAGTGGGGGT-GGCCGAGACGACCCCGCCCGTCTCGAGAGCTCTTCAGA
134270_ENSDARG00000020961_ZEBRAFISH_5965_8474/1-2687 TTTGACAGTTCAGGGCGCGGTCTCGTCCTGGCGCGATCTTAGCTCAATGAAAAGC-AGCC
134270_ENSG00000164853_HUMAN_5374_7839/1-2687 CGGNNNNNNNN-NNNNNNNNNNNNNNNNNATTTTCAACATCAAAATTTCATAATCGCCTC
134270_SINFRUG00000149320_FUGU_3315_5852/1-2687 NNNNNNNNNNN-NNNNNNNNNNNNNNNNNNNNNNN-NNNNNNNNNNNNNNNNNNNNNGTG
134270_ENSRNOG00000001284_RAT_5040_7483/1-2687 GGCCCTCGGGC-TGGCGGTGGCGCGGCGGGGAGTC-ACTTGGCTCCAACTGGAGCGCGCA
134270_ENSMUSG00000029546_MOUSE_5013_7498/1-2687 GGCCCTCGGGC-TGGCAGTGGCGCGGCGGGGAGTC-ACTTGGCTCCAACTGGAGCGCGCA
134270_43-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 5
Columns: 300
Mean pairwise identity: 35.02
Mean single sequence MFE: -80.91
Consensus MFE: -8.17
Energy contribution: -9.70
Covariance contribution: 1.53
Mean z-score: -0.66
Structure conservation index: 0.10
SVM decision value: 0.06
SVM RNA-class probability: 0.561898
Prediction: RNA
######################################################################
>134270_ENSDARG00000020961_ZEBRAFISH_5965_8474/1-2687
AGCACCCCACUGCAUUUCAAUGUUUAAAUUUUAAUGAACCCUUUAAAAAUGUCUCAAGAUGUAGCCUUUUAUCCCUCUAUCUUACAAAAAGUGCUUUUAGAAGUUUGGGAAUCUUUGAAUGUUUUUUUCUUUGUGUCUUAAACUUAUUUUAUCAUACUGUACACAAGUAUGAAUAAUUCGGUUUAUCCAGAUCCCCCUUUUGAACAUCAGAAUUCUAUAAUUGCAGCUUUGACAGUUCAGGGCGCGGUCUCGUCCUGGCGCGAUCUUAGCUCAAUGAAAAGCAGCC
.........((((.((((.((((((((((((..(((((((..(((((((((((....))))).(((((((................))))).))))))))((((((((((......(((.......)))......))))))))))......(((((((.......))))))).......)))))))..)))).......))))))))............((((.((((..((..((((((((((......)))))))).))..))..)))))))))))).)))).. ( -63.70)
>134270_ENSG00000164853_HUMAN_5374_7839/1-2687
CCGCGCCGCGGCGGCUCCGUGCGUCCCAAUGCUCGCAUUUCAAUCGCCCUGUGUUUUUCAUGCUGUCGGGCCGGCUCGCGCUCCGAAAACCUCCUCGAGUGUUUGAGAGGGGCCUUUAAUGACGGUGGGGGCGGCCGGGACGACCCCCGCCCGGCUCGAGAGCUCUUCAGAGGCCCUCGGCUGCGGCGGCGCCGCGGGGAGUCGCUCGGCUUCUGCUCUCGGNNNNNNNNNNNNNNNNNNNNNNNNNAUUUUCAACAUCAAAAUUUCAUAAUCGCCUC
..((((((.(((((((((.((((((((((((....))))....((((((((((.....)))(((((((((((..(((((((((.((........))))))))..)))...))))).....)))))).)))))))..))))).....(((((((((.((((.((((....)).)).)))))))).)))))....))).)))))))))))))..(((....)))...................................................))... ( -106.30)
>134270_SINFRUG00000149320_FUGU_3315_5852/1-2687
AGCAGCCCUCUGUAUUUCAGCGCGUGCAGCCCCACAUUCUAACGAACUCCUCACAAAUCUGCCUUAUUCAUCCUUCCUCCUCUUUUAAGCGCUUCUUUCUUCGGAUUUUGAGGGCGAUCUGAUGAAUAAUUGCCCCUCUCUCGGUCUCUUUUUGAACAUCUAAAAUUCAGUGAUUGCUUUUGGUCAUUUUCAGGCACGACUCCAACCNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGUG
.((.((((((........((((((((......))).(((....)))..........................................)))))....((....))....))))))...(((((((((....(((........))).((.....)).........)))))((((((......))))))..))))))............................................................................... ( -38.60)
>134270_ENSRNOG00000001284_RAT_5040_7483/1-2687
CCGUGCGCCGCAGCCGGCUCCACGCAUCUCAAUGCGACCAUUUCAAUCGCCCAGUGGCNNNNNNNNNNNNNNNNNNNNNNNCUCAUGUGCUCGGGACCCUNNNNNNNNNNNNNNNUCUCAAGCGUUGGAGUGGGGGACUUUAAUGACGGUGGGGGUGGCCGAGUCGACCCCGCCCGGCUCGAGAGCUCUUCAGAGGCCCUCGGGCUGGCGGUGGCGCGGCGGGGAGUCACUUGGCUCCAACUGGAGCGCGCA
...(((((....((((.((((((((((....))))..((((((((((.(((....)))..............................(((.((((...................)))).)))))))))))))...............)))))).))))(((((.((((((((((((((((((.((((....)).)).)))))))))(((....))))))))))..)))))))(((((....)))))))))) ( -100.31)
>134270_ENSMUSG00000029546_MOUSE_5013_7498/1-2687
CCGCGCGCUGCAGCCGGCUCCACGCAUCUCAAUGCGACCAUUUCAAUCGCCCAGUGGCUUUUUNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGUGCCCGGGACCCUNNNNNNNNNNNNNNNNNNCUCAAGCGUUGGAGUGGGGGACUUUAAUGACAGUGGGGGUGGCCGAGACGACCCCGCCCGUCUCGAGAGCUCUUCAGAGGCCCUCGGGCUGGCAGUGGCGCGGCGGGGAGUCACUUGGCUCCAACUGGAGCGCGCA
..(((((((.((((((.(.((((((((....))))(.(((........(((....)))...............................................((((((((((((....................((..(((((((((.....)))))))))....)))))))((((((((((........))))))(((.....)))...))))))))))))))).)))).)))))..((((((....))))))...)).))))))). ( -95.65)
>consensus
__CAGCGCGCUGCAGCCGGCUCCAUGC_AUCUCA_AUG__C___U__ACCAUUUC_AAUCGCCCAGUGUUUU_U__UCCU______________G_GCUC__A______U____GA_UG__UGAGCGGGACCUU___UG____________UC__UCAAG_____CGU__UGGACUCGAGGACUUUAAAGACGGUGGGGGC_GGCCGAGACGACCCCGCCCGGCUCGAGACCUCUGCAGAGGC____G__C__GGC___G_C_CG_C__GGAGUC_ACUUGGCUCCAACAGAAGCGCGCA
....((((((...................................................................................................................((((((.............................................((....)).........((((((..............))))))..))))))..........................................(((((.......))))).......)))))). ( -8.17 = -9.70 + 1.53)
134270_43-rev.rnaz
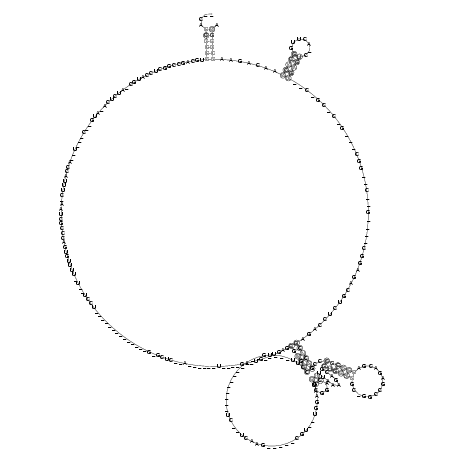
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004